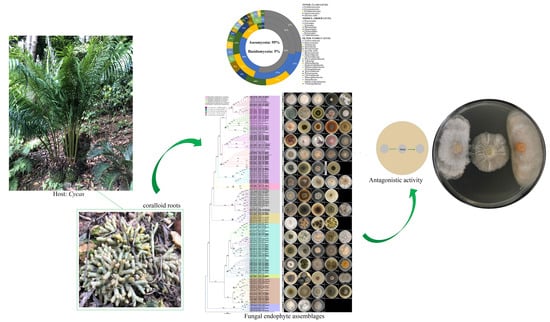
Diversity, Phylogeny and Antagonistic Activity of Fungal Endophytes Associated with Endemic Species of Cycas (Cycadales) in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site Descriptions and Sample Collection
2.2. Coralloid Root Processing and Fungal Endophyte Isolation
2.3. Morphocultural Characterization and Morphotype Groupings of Fungal Endophytes
2.4. DNA Extraction, PCR Amplification and ITS Gene Sequencing
2.5. Molecular Identification and Phylogenetic Analysis of Fungal Endophytes
2.6. Statistical and Diversity Analysis
2.7. In Vitro Co-Cultivation Assay between Fungal Endophytes and the Test Pathogens
3. Results
3.1. Fungal Isolates Characterization and Taxonomic Assignment
3.2. Phylogenetic Analysis of Fungal Endophytes Associated to Cycas debaoensis and Cycas fairylakea
3.3. Diversity and Community Analysis of Fungal Endophytes
3.4. Screening of Antagonistic Potential Using Co-Cultivation Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- dela Cruz, T.E.E.; Notarte, K.I.; Apurillo, C.C.S.; Tarman, K.; Bungihan, M.E. Biomining fungal endophytes from tropical plants and seaweeds for drug discovery. In Biodiversity and Biomedicine; Academic Press: Cambridge, MA, USA, 2020; pp. 51–62. [Google Scholar]
- Mayerhofer, M.S.; Kernaghan, G.; Harper, K. The effects of fungal root endophytes on plant growth: A meta-analysis. Mycorrhiza 2012, 23, 119–128. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of icrobial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- González-Teuber, M.; Vilo, C.; Bascuñán-Godoy, L. Molecular characterization of endophytic fungi associated with the roots of Chenopodium quinoa inhabiting the Atacama Desert, Chile. Genom. Data 2017, 11, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.H.; Radwan, U.; El-Zayat, S.; El-Sayed, M.A. Desert Plant-Fungal Endophytic Association: The Beneficial Aspects to their Hosts. Biol. Forum 2018, 10, 138–145. [Google Scholar]
- Macia-Vicente, J.G.; Jansson, H.-B.; Abdullah, S.K.; Descals, E.; Salinas, J.; Lopez-Llorca, L.V. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol. Ecol. 2008, 64, 90–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciá-Vicente, J.G.; Ferraro, V.; Burruano, S.; Lopez-Llorca, L.V. Fungal Assemblages Associated with Roots of Halophytic and Non-halophytic Plant Species Vary Differentially Along a Salinity Gradient. Microb. Ecol. 2012, 64, 668–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hereme, R.; Morales-Navarro, S.; Ballesteros, G.; Barrera, A.; Ramos, P.; Gundel, P.; Molina-Montenegro, M.A. Fungal Endophytes Exert Positive Effects on Colobanthus quitensis Under Water Stress but Neutral Under a Projected Climate Change Scenario in Antarctica. Front. Microbiol. 2020, 11, 264. [Google Scholar] [CrossRef]
- Zhou, W.-N.; White, J.; Soares, M.; Torres, M.S.; Zhou, Z.-P.; Li, H.-Y. Diversity of fungi associated with plants growing in geothermal ecosystems and evaluation of their capacities to enhance thermotolerance of host plants. J. Plant Interac. 2015, 10, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.Q.; Lan, F.; Qiao, Y.M.; Wei, J.G.; Huang, R.S.; Li, L.B. Endophytic fungi harbored in the root of Sophora tonkinensis Gapnep: Diversity and biocontrol potential against phytopathogens. MicrobiologyOpen 2017, 6, e00437. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, A.R.A. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Macia-Vicente, J.G.; Jansson, H.-B.; Mendgen, K.; Lopez-Llorca, L.V. Colonization of barley roots by endophytic fungi and their reduction of take-all caused by Gaeumannomyces graminis var. tritici. Can. J. Microbiol. 2008, 54, 600–609. [Google Scholar] [CrossRef] [Green Version]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Fang, K.; Miao, Y.-F.; Chen, L.; Zhou, J.; Yang, Z.-P.; Dong, X.-F.; Zhang, H.-B. Tissue-Specific and Geographical Variation in Endophytic Fungi of Ageratina adenophora and Fungal Associations with the Environment. Front. Microbiol. 2019, 10, 2919. [Google Scholar] [CrossRef] [Green Version]
- David, A.S.; Seabloom, E.; May, G. Plant Host Species and Geographic Distance Affect the Structure of Aboveground Fungal Symbiont Communities, and Environmental Filtering Affects Belowground Communities in a Coastal Dune Ecosystem. Microb. Ecol. 2016, 71, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.; Poudel, R.; Nebel, K.A.; Collins, S.L. Precipitation increases the abundance of some groups of root-associated fungal endophytes in a semiarid grassland. Ecosphere 2011, 2, 50. [Google Scholar] [CrossRef]
- Gallart, M.; Adair, K.L.; Love, J.; Meason, D.; Clinton, P.; Xue, J.; Turnbull, M.H. Host Genotype and Nitrogen form Shape the Root Microbiome of Pinus radiata. Microb. Ecol. 2018, 75, 419–433. [Google Scholar] [CrossRef]
- Qian, X.; Duan, T.; Sun, X.; Zheng, Y.; Wang, Y.; Hu, M.; Yao, H.; Ji, N.; Lv, P.; Chen, L.; et al. Host genotype strongly influences phyllosphere fungal communities associated with Mussaenda pubescens var. alba (Rubiaceae). Fungal Ecol. 2018, 36, 141–151. [Google Scholar] [CrossRef]
- Notarte, K.; Nakao, Y.; Yaguchi, T.; Bungihan, M.; Suganuma, K.; dela Cruz, T.E.E. Trypanocidal activity, cytotoxicity and histone modifications induced by malformin A1 isolated from the marine-derived fungus Aspergillus tubingensis IFM 63452. Mycosphere 2017, 8, 111–120. [Google Scholar] [CrossRef]
- Notarte, K.I.; Devanadera, M.K.; Mayor, A.B.; Cada, M.C.; Pecundo, M.H.; Macabeo, A.P. Toxicity, antibacterial, and antioxidant activities of fungal endophytes Colletotrichum and Nigrospora spp. isolated from Uvaria grandiflora. Philipp. J. Sci. 2019, 148, 503–510. [Google Scholar]
- Notarte, K.I.; Yaguchi, T.; Suganuma, K.; dela Cruz, T.E.E. Antibacterial, cytotoxic and trypanocidal activities of marine-derived fungi isolated from Philippine macroalgae and seagrasses. Acta Bot. Croat. 2018, 77, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, C.S.P.; Notarte, K.I.R.; dela Cruz, T.E.E. Antibacterial activities of mangrove leaf endophytic fungi from Luzon Island, Philippines. Studies in Fungi 2020, 3, 320–331. [Google Scholar] [CrossRef]
- Schmeda-Hirschmann, G.; Hormazábal, E.; Astudillo, L.; Rodriguez, J.; Theoduloz, C. Secondary metabolites from endophytic fungi isolated from the Chilean gymnosperm Prumnopitys andina (Lleuque). World J. Microbiol. Biotechnol. 2005, 21, 27–32. [Google Scholar] [CrossRef]
- Hormazábal, E.; Piontelli, E. Endophytic fungi from Chilean native gymnosperms: Antimicrobial activity against human and phytopathogenic fungi. World J. Microbiol. Biotechnol. 2009, 25, 813–819. [Google Scholar] [CrossRef]
- Duan, X.; Xu, F.; Qin, D.; Gao, T.; Shen, W.; Zuo, S.; Yu, B.; Xu, J.; Peng, Y.; Dong, J. Diversity and bioactivities of fungal endophytes from Distylium chinense, a rare waterlogging tolerant plant endemic to the Three Gorges Reservoir. BMC Microbiol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.L.; Guo, T.T.; Ren, Z.X.; Zhang, N.S.; Wang, M.L. Diversity and antioxidant activity of culturable endophytic fungi from alpine plants of Rhodiola crenulata, R. angusta, and R. sachalinensis. PLoS ONE 2015, 10, e0118204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.-M.; Zhou, Y.-Q.; Zhou, X.-L.; Xia, X.-H.; Wei, Y.; He, L.-L.; Tang, H.-Z.; Yu, L.-Y. Diversity and bioactive potential of culturable fungal endophytes of Dysosma versipellis; a rare medicinal plant endemic to China. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkateswarulu, N.; Shameer, S.; Bramhachari, P.; Basha, S.T.; Nagaraju, C.; Vijaya, T. Isolation and characterization of plumbagin (5- hydroxyl- 2- methylnaptalene-1,4-dione) producing endophytic fungi Cladosporium delicatulum from endemic medicinal plants. Biotechnol. Rep. 2018, 20, e00282. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.C.; Chen, C.J. Cycas debaoensis Y. C. Zhong & C. J. Chen—A New Cycad from China. Acta Phytotax. 1997, 35, 571. [Google Scholar]
- Wang, D.Y. Taxonomy of Cycas in China. In Cycads in China; Wang, F.X., Liang, H.B., Chen, T.Q., Wang, D.Y., Eds.; Guangdong Science and Technology Press: Guangdong, China, 1996; pp. 13–19. [Google Scholar]
- Chang, A.C.G.; Chen, T.; Li, N.; Duan, J. Perspectives on Endosymbiosis in Coralloid Roots: Association of Cycads and Cyanobacteria. Front. Microbiol. 2019, 10, 1888. [Google Scholar] [CrossRef] [Green Version]
- Jian, S.; Liu, N.; Gao, Z.; Wei, Q.; Xie, Z.; Wu, M.; Ren, H. Biological characteristics of wild Cycas fairylakea population in Guangdong Province, China. Front. Biol. China 2006, 1, 430–433. [Google Scholar] [CrossRef]
- Jianguang, X.; Shuguang, J.; Nian, L. Genetic variation in the endemic plant Cycas debaoens is on the basis of ISSR analysis. Aust. J. Bot. 2005, 53, 141–145. [Google Scholar] [CrossRef]
- Xiao, L.; Ge, X.; Gong, X.; Hao, G.; Zheng, S. ISSR Variation in the Endemic and Endangered Plant Cycas guizhouensis (Cycadaceae). Ann. Bot. 2004, 94, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-García, K.; Bustos-Díaz, E.D.; Corona-Gomez, J.A.; Ramos-Aboites, H.; Selem, N.; Cruz-Morales, P.; Pérez-Farrera, M.; Barona-Gómez, F.; Cibrián-Jaramillo, A. Cycad Coralloid Roots Contain Bacterial Communities Including Cyanobacteria and Caulobacter spp. That Encode Niche-Specific Biosynthetic Gene Clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Chiang, T.-Y.; Huang, C.-L.; Gong, X. Highly diverse endophytes in roots of Cycas bifida (Cycadaceae), an ancient but endangered gymnosperm. J. Microbiol. 2018, 56, 337–345. [Google Scholar] [CrossRef]
- Zheng, Y.; Gong, X. Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome 2019, 7, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Pecundo, M.H.; Chang, A.C.G.; Chen, T.; dela Cruz, T.E.E.; Ren, H.; Li, N. Full-Length 16S rRNA and ITS Gene Sequencing Revealed Rich Microbial Flora in Roots of Cycas spp. in China. Evol. Bioinform. 2021, 17, 1–16. [Google Scholar] [CrossRef]
- Chang, A.C.G.; Pecundo, M.H.; Duan, J.; Ren, H.; Chen, T.; Li, N. GeoChip 5.0 microarray: A descriptive overview of genes present in coralloid root microbiomes of Cycas debaoensis and Cycas fairylakea. ELBA Bioflux 2020, 12, 5–20. [Google Scholar]
- Solis, M.J.L.; dela Cruz, T.E.E.; Schnittler, M.; Unterseher, M. The diverse community of leaf-inhabiting fungal endophytes from Philippine natural forests reflects phylogenetic patterns of their host plant species Ficus benjamina, F. elastica and F. religiosa. Mycoscience 2016, 57, 96–106. [Google Scholar] [CrossRef]
- Aamir, S. A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol. Quar. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Solis, M.J.L.; Yurkov, A.; dela Cruz, T.E.E.; Unterseher, M. Leaf-inhabiting endophytic yeasts are abundant but unevenly distributed in three Ficus species from botanical garden greenhouses in Germany. Mycol. Prog. 2014, 14, 1019. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Dagamac, N.H.A.; Sogono, P.G.; Cabalfin, R.C.B.; Adducul, A.C.Y.; dela Cruz, T.E.E. Fungal Root Endophytes from Musa spp. as Biological Control Agents against the Plant Pathogen Fusarium oxysporum. Acta Manil. 2010, 56, 27–35. [Google Scholar] [CrossRef]
- De Mesa, R.B.C.; Espinosa, I.R.; Agcaoili, M.C.R.R.; Calderon, M.A.T.; Pangilinan, M.V.B.; De Padua, J.C.; dela Cruz, T.E.E. Antagonistic activities of needle-leaf fungal endophytes against Fusarium spp. MycoAsia 2020, 6, 1–11. [Google Scholar]
- dela Cruz, T.E.E.; Din, H.J.F.; Aril-dela Cruz, J.V. Microbes for Sustainable Agriculture: Isolation and Identification of Beneficial Soil- and Plant-Associated Microorganisms. In SEARCA Professorial Chair Lecture Monograph No. 6; SEARCA: Laguna, Philippines, 2021. [Google Scholar]
- Hamzah, T.N.T.; Lee, S.Y.; Hidayat, A.; Terhem, R.; Faridah-Hanum, I.; Mohamed, R. Diversity and Characterization of Endophytic Fungi Isolated from the Tropical Mangrove Species, Rhizophora mucronata, and Identification of Potential Antagonists Against the Soil-Borne Fungus, Fusarium solani. Front. Microbiol. 2018, 9, 1707. [Google Scholar] [CrossRef]
- Wheeler, K.A.; Hocking, A.D. Interactions among xerophilic fungi associated with dried salted fish. J. Appl. Bacteriol. 1993, 74, 164–169. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, B.; Yu, Z.; Jiang, S.; Yang, Y. Study on the Diversity of Endophytic Fungi in Cycas panzhihuaensis L. J. Life Sci. Res. 2013, 17, 406–411. [Google Scholar]
- Apurillo, C.C.S.; Cai, L.; dela Cruz, T.E.E. Diversity and bioactivities of mangrove fungal endophytes from Leyte and Samar, Philippines. Philipp. Sci. Lett. 2019, 12, 33–48. [Google Scholar]
- Chi, F.; Shen, S.-H.; Cheng, H.-P.; Jing, Y.-X.; Yanni, Y.G.; Dazzo, F.B. Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, inside Rice Plants and Assessment of Benefits to Rice Growth Physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [Green Version]
- Hodgson, S.; De Cates, C.; Hodgson, J.; Morley, N.J.; Sutton, B.C.; Gange, A.C. Vertical transmission of fungal endophytes is widespread in forbs. Ecol. Evol. 2014, 4, 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.-K.; Miao, C.-P.; Chen, H.-H.; Huang, F.-F.; Xia, Y.-M.; Chen, Y.-W.; Zhao, L.-X. Endophytic fungi harbored in Panax notoginseng: Diversity and potential as biological control agents against host plant pathogens of root-rot disease. J. Ginseng Res. 2017, 41, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Doilom, M. Can ITS sequence data identify fungal endophytes from cultures? A case study from Rhizophora apiculata. Mycosphere 2017, 8, 1869–1892. [Google Scholar] [CrossRef]
- Jumpponen, A.; Herrera, J.; Porras-Alfaro, A.; Rudgers, J. Biogeography of root-associated fungal endophytes. In Biogeography of Mycorrhizal Symbiosis; Tedersoo, L., Ed.; Springer: Cham, FL, USA, 2017; pp. 195–222. [Google Scholar]
- Knapp, D.G.; Imrefi, I.; Boldpurev, E.; Csíkos, S.; Akhmetova, G.; Berek-Nagy, P.J.; Otgonsuren, B.; Kovács, G.M. Root-Colonizing Endophytic Fungi of the Dominant Grass Stipa krylovii From a Mongolian Steppe Grassland. Front. Microbiol. 2019, 10, 2565. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.M.N.; Haque, M.A.; Chowdhury, M.Z.H.; Mishu, N.J.; Hossain, M.M. A Hypocrealean Fungus Penicillifer diparietisporus, First Reported in Bangladesh. Bangladesh J. Sci. Res. 2020, 31–33, 63–65. [Google Scholar]
- Das, K.; Back, C.G.; Lee, S.Y.; Jung, H.Y. Penicillifer diparietisporus: A New Record from Field Soil in Korea. Korean J. Mycol. 2019, 46. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Zhang, G.; Fu, C.; Liu, M.; Xing, Q.; et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Pither, J.; Pickles, B.J. The paleosymbiosis hypothesis: Host plants can be colonized by root symbionts that have been inactive for centuries to millenia. FEMS Microbiol. Ecol. 2017, 93, 1–9. [Google Scholar] [CrossRef]
- Li, Y.-B.; Shao, J.-A.; Yang, H.; Bai, X. The relations between land use and karst rocky desertification in a typical karst area, China. Environ. Earth Sci. 2008, 57, 621–627. [Google Scholar] [CrossRef]
- Tian, Q.; Taniguchi, T.; Shi, W.-Y.; Li, G.; Yamanaka, N.; Du, S. Land-use types and soil chemical properties influence soil microbial communities in the semiarid Loess Plateau region in China. Sci. Rep. 2017, 7, srep45289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nat. Cell Biol. 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Latz, M.A.; Kerrn, M.H.; Sørensen, H.; Collinge, D.B.; Jensen, B.; Brown, J.K.; Madsen, A.M.; Jørgensen, H.J.L. Succession of the fungal endophytic microbiome of wheat is dependent on tissue-specific interactions between host genotype and environment. Sci. Total Environ. 2021, 759, 143804. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.A.; Jumpponen, A.; Rudgers, J.A. Divergence in Diversity and Composition of Root-Associated Fungi Between Greenhouse and Field Studies in a Semiarid Grassland. Microb. Ecol. 2018, 78, 122–135. [Google Scholar] [CrossRef]
- Schöps, R.; Goldmann, K.; Herz, K.; Lentendu, G.; Schöning, I.; Bruelheide, H.; Wubet, T.; Buscot, F. Land-Use Intensity Rather Than Plant Functional Identity Shapes Bacterial and Fungal Rhizosphere Communities. Front. Microbiol. 2018, 9, 2711. [Google Scholar] [CrossRef]
- Schöps, R.; Goldmann, K.; Korell, L.; Bruelheide, H.; Wubet, T.; Buscot, F. Resident and phytometer plants host comparable rhizosphere fungal communities in managed grassland ecosystems. Sci. Rep. 2020, 10, 919. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, L. Diversity of Colletotrichum Species Associated with Anthracnose Disease in Tropical Fruit Crops—A Review. Agriculture 2021, 11, 297. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Dissanayake, A.J.; Li, X.; Liu, M.; Wanasinghe, D.; Xu, J.; Zhao, W.; Zhang, W.; Zhou, Y.; Hyde, K.D.; et al. High Genetic Diversity and Species Complexity of Diaporthe Associated with Grapevine Dieback in China. Front. Microbiol. 2019, 10, 1936. [Google Scholar] [CrossRef]
- Vidal, A.; Parada, R.; Mendoza, L.; Cotoras, M. Endophytic Fungi Isolated from Plants Growing in Central Andean Precordillera of Chile with Antifungal Activity against Botrytis cinerea. J. Fungi 2020, 6, 149. [Google Scholar] [CrossRef]
- You, F.; Han, T.; Wu, J.-Z.; Huang, B.-K.; Qin, L.-P. Antifungal secondary metabolites from endophytic Verticillium sp. Biochem. Syst. Ecol. 2009, 37, 162–165. [Google Scholar] [CrossRef]
- Gharsallah, H.; Ksentini, I.; Naayma, S.; Taieb, K.H.; Abdelhedi, N.; Schuster, C.; Triki, M.A.; Ksantini, M.; Leclerque, A. Identification of fungi in Tunisian olive orchards: Characterization and biological control potential. BMC Microbiol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.R.; Edwards, R.L.; Whalley, A.J.S. Metabolites of the higher fungi. Part 21. 3-Methyl-3,4-dihydroisocoumarins and related compounds from the ascomycete family xylariaceae. J. Chem. Soc. Perkin Trans. 1983, 1, 2185–2192. [Google Scholar] [CrossRef]
- Inose, K.; Tanaka, K.; Koshino, H.; Hashimoto, M. Cyclopericodiol and new chlorinated melleins isolated from Periconia macrospinosa KT3863. Tetrahedron 2019, 75, 130470. [Google Scholar] [CrossRef]
- Sun, Z.-B.; Li, S.-D.; Ren, Q.; Xu, J.-L.; Lu, X.; Sun, M.-H. Biology and applications of Clonostachys rosea. J. Appl. Microbiol. 2020, 129, 486–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.-B.; Sun, M.-H.; Li, S.-D. Draft Genome Sequence of Mycoparasite Clonostachys rosea Strain 67-1. Genome Announc. 2015, 3, e00546-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Ren, J.; Li, H.; Pan, Y.; Liu, X.; Che, Y.; Liu, G. The disruption of verM activates the production of gliocladiosin A and B in Clonostachys rogersoniana. Org. Biomol. Chem. 2019, 17, 6782–6785. [Google Scholar] [CrossRef]
- Pombeiro-Sponchiado, S.R.; Sousa, G.S.; Andrade, J.C.R.; Lisboa, H.F.; Gonçalves, R.C.R. Production of Melanin Pigment by Fungi and Its Biotechnological Applications. In Melanin; IntechOpen: London, UK, 2017. [Google Scholar]
- Davidson, J.F.; Whyte, B.; Bissinger, P.H.; Schiestl, R.H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5116–5121. [Google Scholar] [CrossRef] [Green Version]
- Redman, R.S.; Vervoort, V.S.; Beachem, M.A.; Edwards, P.S.; Ladd, S.; Miller, K.E.; De Mollerat, X.; Clarkson, K.; Dupont, B.; Schwartz, C.E.; et al. Thermotolerance Generated by Plant/Fungal Symbiosis. Science 2002, 298, 1581. [Google Scholar] [CrossRef]






| Host | Population Sites | Locality | GPS/Altitude | Habitat Description | Collection Date |
|---|---|---|---|---|---|
| Cycas debaoensis | botanical garden | 1 NCGCC, FLBG, Shenzhen, Guangdong | 22°34′36.1″ N 114°10′51.1″ E 60 ± 30 m.a.s.l. | Living collections of cycads in a controlled and managed environment; individuals used for the study were planted in 2004. | 26 January 2020 |
| natural habitat | Debao County, Baise, Guangxi | 23°29′29.2″ N 106°12′50.8″ E 905 ± 24 m.a.s.l. | Dispersed in open landscapes and on limestone hills with shrubs and trees; some cycad plants growing on cracks or in thin soil near the edges of limestone. | 22 October 2020 | |
| reintroduction site | 1 Huanglian mountain nature reserve, Debao County, Baise, Guangxi | 23°33′47.1″ N 106°14′09.8″ E 885 ± 8 m.a.s.l. | Scattered on open slopes with grasses and sparsely covered by shrubs and small trees; local population was reintroduced in 2008. | 22 October 2020 | |
| Cycas fairylakea | Botanical garden | 1 NCGCC, FLBG, Shenzhen, Guangdong | 22°34′36.4″ N 114°10′50.6″ E 55 ± 20 m.a.s.l. | Living collections of cycads in controlled and managed environment; individuals used for the study were planted in 2017. | 26 January 2020 |
| natural habitat | Meilin Reservoir Park, Shenzhen, Guangdong | 22°34′38.5″ N 114°00′34.2″ E 114 ± 23 m.a.s.l. | Growing on slopes of ridges and cliffs along river valleys and understory of moist closed subtropical forests at low elevations. | 21 January 2020 |
| 1 Sample Source | 2 Records of Fungal Endophytes | |||||||
|---|---|---|---|---|---|---|---|---|
| R | G | S | S/G Ratio | H’ | FA | CR (%) | IF (%) | |
| Cdeb, Nh | 37 | 12 | 14 | 1.2 a | 2.2 a | 8.2 a | 17 | 13 |
| Cfai, Nh | 52 | 16 | 18 | 1.1 a | 2.2 a | 9.8 b | 24 | 18 |
| Cdeb, Bg | 77 | 15 | 23 | 1.5 b | 2.7 b,c | 11.1 c | 36 | 27 |
| Cfai, Bg | 74 | 15 | 21 | 1.4 a,b | 2.5 c | 9.8 b | 34 | 26 |
| Cdeb, Re | 44 | 13 | 15 | 1.2 a | 2.2 a | 8.0 a | 20 | 15 |
| Cdeb (Pooled) | 158 | 29 | 41 | 1.4 a | 3.0 a | 17.8 a | 24 | 56 |
| Cfai (Pooled) | 126 | 21 | 32 | 1.5 a | 2.7 b | 13.8 b | 29 | 44 |
| Species Name and Isolate Code | Clade | Diaporthe sp. Strain CdP01 | Colletotrichum sp. Strain CdP02 | ||
|---|---|---|---|---|---|
| Percent Inhibition (I%) ± SD | Type of Interaction | Percent Inhibition (I%) ± SD | Type of Interaction | ||
| Castanediella sp. GD016 | G | 58.7 ± 1.89 | C | 63.3 ± 1.57 | C |
| Clonostachys rogersoniana BD005 | E | 52 ± 1.63 | B | 62.2 ± 1.50 | C |
| Clonostachys rogersoniana BF024 | E | 66.7 ± 1.89 | F | 82.1 ± 1.50 | F |
| Clonostachys rosea BF011 | E | 64.3 ± 2.05 | F | 80 ± 1.63 | F |
| Curvularia sp. BF002 | M | 50 ± 1.63 | B | 56.7 ± 2.72 | B |
| Curvularia sp. GD014 | M | 51.6 ± 0.47 | B | 53.9 ± 2.1 | B |
| Gliocladiopsis sp. GD004 | C | 56 ± 3.27 | B | 68.3 ± 1.36 | E |
| Hypoxylon sp. GD019 | G | 87.33 ± 0.94 | C | 70.3 ± 1.89 | C |
| Ilyonectria sp. GD020 | C | 60 ± 3.77 | B | 61.7 ± 1.70 | B |
| Microsphaeropsis arundinis BF008 | N | 42.7 ± 3.77 | B | 51.1 ± 2.83 | B |
| Penicillifer sp. GD008 | C | 69.3 ± 3.30 | F | 81.1 ± 3.14 | F |
| Penicillium sp. BD022 | J | 64 ± 2.83 | C | 81.5 ± 2.51 | C |
| Periconia sp. GD026 | O | 62 ± 2.83 | C | 69.3 ± 1.89 | C |
| Phoma sp. GD023 | M | 61. 3 ± 4.99 | B | 65.6 ± 4.16 | B |
| Pseudopithomyces maydicus GD018 | N | 58.7 ± 1.89 | B | 65.6 ± 1.57 | B |
| Alternaria/Setophoma sp. GD005 | M | 61.3 ± 1.89 | C | 75.6 ± 1.57 | C |
| Alternaria/Setophoma sp. BD019 | M | 62. 3 ± 0.94 | C | 71.1 ± 2.83 | C |
| Verticillium sp. MF026 | A | 53.3 ± 1.89 | C | 65.56 ± 1.57 | C |
| Verticillium sp. GD030 | A | 49.3 ± 1.89 | E | 58 ± 2.79 | E |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecundo, M.H.; dela Cruz, T.E.E.; Chen, T.; Notarte, K.I.; Ren, H.; Li, N. Diversity, Phylogeny and Antagonistic Activity of Fungal Endophytes Associated with Endemic Species of Cycas (Cycadales) in China. J. Fungi 2021, 7, 572. https://doi.org/10.3390/jof7070572
Pecundo MH, dela Cruz TEE, Chen T, Notarte KI, Ren H, Li N. Diversity, Phylogeny and Antagonistic Activity of Fungal Endophytes Associated with Endemic Species of Cycas (Cycadales) in China. Journal of Fungi. 2021; 7(7):572. https://doi.org/10.3390/jof7070572
Chicago/Turabian StylePecundo, Melissa H., Thomas Edison E. dela Cruz, Tao Chen, Kin Israel Notarte, Hai Ren, and Nan Li. 2021. "Diversity, Phylogeny and Antagonistic Activity of Fungal Endophytes Associated with Endemic Species of Cycas (Cycadales) in China" Journal of Fungi 7, no. 7: 572. https://doi.org/10.3390/jof7070572