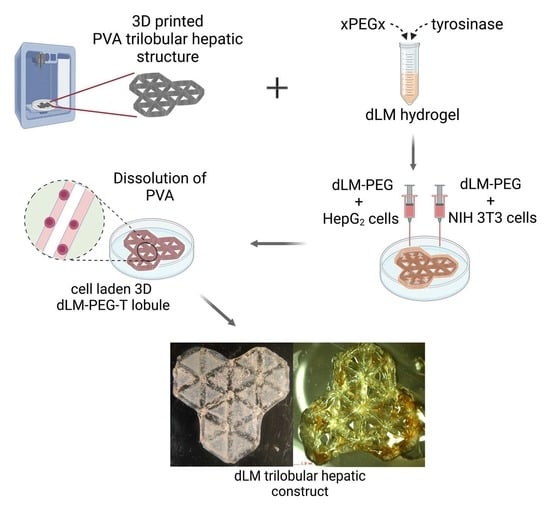
Indirect 3D Bioprinting of a Robust Trilobular Hepatic Construct with Decellularized Liver Matrix Hydrogel
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. 3D Printing of PVA
2.3. Preparation of dLM-PEG-T Hydrogel
2.4. Rheology and Degree of Crosslinking
2.5. Cell Culture and Encapsulation in dLM-PEG Scaffold
2.6. Preparation of Cell-Laden dLM-PEG-T Trilobular 3D Construct
2.7. Cell Viability, Proliferation, and Metabolic Activity
2.8. Functional Assessment and Gene Expression Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Preparation and Characterization of dLM-PEG-T Hydrogel
3.2. Development of 3D Trilobular Hepatic Construct with PVA and dLM-PEG-T
3.3. Cell Viability and Proliferation within the dLM-PEG-T Construct
3.4. Liver-Specific Functions and Gene Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vrana, N.E.; Gupta, S.; Mitra, K.; Rizvanov, A.A.; Solovyeva, V.V.; Antmen, E.; Salehi, M.; Ehterami, A.; Pourchet, L.; Barthes, J.; et al. From 3D printing to 3D bioprinting: The material properties of polymeric material and its derived bioink for achieving tissue specific architectures. Cell Tissue Bank. 2022, 23, 417–440. [Google Scholar] [CrossRef] [PubMed]
- Leucht, A.; Volz, A.-C.; Rogal, J.; Borchers, K.; Kluger, P.J. Advanced gelatin-based vascularization bioinks for extrusion-based bioprinting of vascularized bone equivalents. Sci. Rep. 2020, 10, 5330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklanny, A.A.; Machour, M.; Redenski, I.; Chochola, V.; Goldfracht, I.; Kaplan, B.; Epshtein, M.; Yameen, H.S.; Merdler, U.; Feinberg, A.; et al. 3D bioprinting of engineered tissue flaps with hierarchical vessel networks (vesselnet) for direct host-to-implant perfusion. Adv. Mater. 2021, 33, 2102661. [Google Scholar] [CrossRef] [PubMed]
- Skylar-Scott, M.A.; Uzel, S.G.M.; Nam, L.L.; Ahrens, J.H.; Truby, R.L.; Damaraju, S.; Lewis, J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019, 5, eaaw2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolesky, D.B.; Homan, K.A.; Skylar-Scott, M.A.; Lewis, J.A. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. USA 2016, 113, 3179–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavafoghi, M.; Darabi, M.A.; Mahmoodi, M.; Tutar, R.; Xu, C.; Mirjafari, A.; Billi, F.; Swieszkowski, W.; Nasrollahi, F.; Ahadian, S.; et al. Multimaterial bioprinting and combination of processing techniques towards the fabrication of biomimetic tissues and organs. Biofabrication 2021, 13, 042002. [Google Scholar] [CrossRef]
- Pati, F.; Jang, J.; Ha, D.-H.; Kim, S.W.; Rhie, J.-W.; Shim, J.-H.; Kim, D.-H.; Cho, D.-W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014, 5, 3935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Cho, D.-W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 2016, 16, 2618–2625. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Derakhshanfar, S.; Zhong, W.; Li, B.; Lu, F.; Xing, M.; Li, X. Characterization and application of carboxymethyl chitosan-based bioink in cartilage tissue engineering. J. Nanomater. 2020, 2020, 2057097. [Google Scholar] [CrossRef]
- Diamantides, N.; Wang, L.; Pruiksma, T.; Siemiatkoski, J.; Dugopolski, C.; Shortkroff, S.; Kennedy, S.; Bonassar, L.J. Correlating rheological properties and printability of collagen bioinks: The effects of riboflavin photocrosslinking and pH. Biofabrication 2017, 9, 034102. [Google Scholar] [CrossRef]
- Houben, A.; Van Hoorick, J.; Van Erps, J.; Thienpont, H.; Van Vlierberghe, S.; Dubruel, P. Indirect rapid prototyping: Opening up unprecedented opportunities in Scaffold design and applications. Ann. Biomed. Eng. 2016, 45, 58–83. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Hudson, A.R.; Shiwarski, D.J.; Tashman, J.W.; Hinton, T.J.; Yerneni, S.; Bliley, J.M.; Campbell, P.G.; Feinberg, A.W. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019, 365, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.E.; Duchi, S.; Onofrillo, C.; Quigley, A.; Di Bella, C.; Pirogova, E.; O’Connell, C.D. Printing between the lines: Intricate biomaterial structures fabricated via negative embodied sacrificial template 3D (NEST3D) printing. Adv. Mater. Technol. 2021, 6, 2100189. [Google Scholar] [CrossRef]
- Shao, L.; Gao, Q.; Xie, C.; Fu, J.; Xiang, M.; Liu, Z.; Xiang, L.; He, Y. Sacrificial microgel-laden bioink-enabled 3D bioprinting of mesoscale pore networks. Bio-Design Manuf. 2020, 3, 30–39. [Google Scholar] [CrossRef]
- Tamjid, E.; Simchi, A. Fabrication of a highly ordered hierarchically designed porous nanocomposite via indirect 3D printing: Mechanical properties and in vitro cell responses. Mater. Des. 2015, 88, 924–931. [Google Scholar] [CrossRef]
- Van Hoorick, J.; Declercq, H.; De Muynck, A.; Houben, A.; Van Hoorebeke, L.; Cornelissen, R.; Van Erps, J.; Thienpont, H.; Dubruel, P.; Van Vlierberghe, S. Indirect additive manufacturing as an elegant tool for the production of self-supporting low density gelatin scaffolds. J. Mater. Sci. Mater. Electron. 2015, 26, 247. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Choi, B.; Wu, B.; Lee, M. Customized biomimetic scaffolds created by indirect three-dimensional printing for tissue engineering. Biofabrication 2013, 5, 045003. [Google Scholar] [CrossRef]
- Ze, Y.; Li, Y.; Huang, L.; Shi, Y.; Li, P.; Gong, P.; Lin, J.; Yao, Y. Biodegradable inks in indirect three-dimensional bioprinting for tissue vascularization. Front. Bioeng. Biotechnol. 2022, 10, 461. [Google Scholar] [CrossRef]
- Naghieh, S.; Sarker, M.; Abelseth, E.; Chen, X. Indirect 3D bioprinting and characterization of alginate scaffolds for potential nerve tissue engineering applications. J. Mech. Behav. Biomed. Mater. 2019, 93, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Schöneberg, J.; De Lorenzi, F.; Theek, B.; Blaeser, A.; Rommel, D.; Kuehne, A.J.C.; Kießling, F.; Fischer, H. Engineering biofunctional in vitro vessel models using a multilayer bioprinting technique. Sci. Rep. 2018, 8, 10430. [Google Scholar] [CrossRef]
- Muller, M.; Becher, J.; Schnabelrauch, M.; Zenobi-Wong, M. Printing thermoresponsive reverse molds for the creation of patterned two-component hydrogels for 3D cell culture. J. Vis. Exp. 2013, 77, e50632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchamp, M.; Liu, T.; Van Genderen, A.M.; Kappings, V.; Oklu, R.; Ellisen, L.W.; Zhang, Y.S. Sacrificial bioprinting of a mammary ductal carcinoma model. Biotechnol. J. 2019, 14, e1700703. [Google Scholar] [CrossRef] [PubMed]
- Visser, J.; Peters, B.; Burger, T.J.; Boomstra, J.; Dhert, W.; Melchels, F.P.W.; Malda, J. Biofabrication of multi-material anatomically shaped tissue constructs. Biofabrication 2013, 5, 035007. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, C.R.; Ko, S.K.; Caviglia, C.; Wolff, A.; Emnéus, J.; Keller, S.S.; Dufva, M. Three-dimensional fabrication of thick and densely populated soft constructs with complex and actively perfused channel network. Acta Biomater. 2018, 65, 174–184. [Google Scholar] [CrossRef]
- Miller, J.S.; Stevens, K.R.; Yang, M.T.; Baker, B.M.; Nguyen, D.-H.T.; Cohen, D.M.; Toro, E.; Chen, A.A.; Galie, P.A.; Yu, X.; et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef]
- Bertassoni, L.E.; Cardoso, J.C.; Manoharan, V.; Cristino, A.L.; Bhise, N.S.; Araujo, W.A.; Zorlutuna, P.; Vrana, N.E.; Ghaemmaghami, A.M.; Dokmeci, M.R.; et al. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication 2014, 6, 024105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norotte, C.; Marga, F.S.; Niklason, L.E.; Forgacs, G. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials 2009, 30, 5910–5917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertassoni, L.E.; Cecconi, M.; Manoharan, V.; Nikkhah, M.; Hjortnaes, J.; Cristino, A.L.; Barabaschi, G.; Demarchi, D.; Dokmeci, M.R.; Yang, Y.; et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip 2014, 14, 2202–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolesky, D.B.; Truby, R.L.; Gladman, A.S.; Busbee, T.A.; Homan, K.A.; Lewis, J.A. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 2014, 26, 3124–3130. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, W.; Liu, H.; Chen, Y.; Li, B.; Gou, Z.; Li, X.; Gou, M. 3D printing of functional nerve guide conduits. Burn. Trauma 2021, 9, tkab011. [Google Scholar] [CrossRef]
- Hauser, P.V.; Chang, H.-M.; Nishikawa, M.; Kimura, H.; Yanagawa, N.; Hamon, M. Bioprinting scaffolds for vascular tissues and tissue vascularization. Bioengineering 2021, 8, 178. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Larsen, L.B.; Trifol, J.; Szabo, P.; Burri, H.V.R.; Canali, C.; Dufva, M.; Emnéus, J.; Wolff, A. Fabrication of scalable and structured tissue engineering scaffolds using water dissolvable sacrificial 3D printed moulds. Mater. Sci. Eng. C 2015, 55, 569–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stocco, E.; Barbon, S.; Dalzoppo, D.; Lora, S.; Sartore, L.; Folin, M.; Parnigotto, P.P.; Grandi, C. Tailored PVA/ECM scaffolds for cartilage regeneration. BioMed. Res. Int. 2014, 2014, 762189. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yang, G.H.; Choi, C.H.; Cho, Y.S.; Kim, G. Gelatin/PVA scaffolds fabricated using a 3D-printing process employed with a low-temperature plate for hard tissue regeneration: Fabrication and characterizations. Int. J. Biol. Macromol. 2018, 120, 119–127. [Google Scholar] [CrossRef]
- Dahlberg, T.; Stangner, T.; Zhang, H.; Wiklund, K.; Lundberg, P.; Edman, L.; Andersson, M. 3D printed water-soluble scaffolds for rapid production of PDMS micro-fluidic flow chambers. Sci. Rep. 2018, 8, 3372. [Google Scholar] [CrossRef] [Green Version]
- Zeußel, L.; Hampl, J.; Weise, F.; Singh, S.; Schober, A. Bio-inspired 3D micro structuring of a liver lobule via direct laser writing: A comparative study with SU-8 and SUEX. J. Laser Appl. 2022, 34, 012007. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Balis, U.J.; Yarmush, M.L.; Toner, M. Effect of cell–cell interactions in preservation of cellular phenotype: Cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999, 13, 1883–1900. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-S.; Harn, H.-J.; Hsieh, D.-K.; Wen, T.-C.; Subeq, Y.-M.; Sun, L.-Y.; Lin, S.-Z.; Chiou, T.-W. Cells and materials for liver tissue engineering. Cell Transplant. 2013, 22, 685–700. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Underhill, G.H.; Zaret, K.S.; Fox, I.J. Cell and tissue engineering for liver disease. Sci. Transl. Med. 2014, 6, 245sr2. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, X.; Zhang, L.; Sun, L.; Wang, H.; Zhao, H.; Zhang, Z.; Liu, W.; Huang, Y.; Ji, S.; et al. 3D liver tissue model with branched vascular networks by multimaterial bioprinting. Adv. Health Mater. 2021, 10, 2101405. [Google Scholar] [CrossRef]
- Rowe, C.; Gerrard, D.T.; Jenkins, R.; Berry, A.; Durkin, K.; Sundstrom, L.; Goldring, C.E.; Park, B.K.; Kitteringham, N.R.; Hanley, K.P.; et al. Proteome-wide analyses of human hepatocytes during differentiation and dedifferentiation. Hepatology 2013, 58, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-derived decellularized extracellular matrix: A game changer for bioink manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef] [PubMed]
- Khati, V.; Ramachandraiah, H.; Pati, F.; Svahn, H.A.; Gaudenzi, G.; Russom, A. 3D bioprinting of multi-material decellularized liver matrix hydrogel at physiological temperatures. Biosensors 2022, 12, 521. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, R.; Islam, M.M.; Alarcon, E.I.; Samanta, A.; Wang, S.; Lundström, P.; Hilborn, J.; Griffith, M.; Phopase, J. Functionalised type-I collagen as a hydrogel building block for bio-orthogonal tissue engineering applications. J. Mater. Chem. B 2015, 4, 318–326. [Google Scholar] [CrossRef] [Green Version]
- van Genderen, A.M.; Valverde, M.G.; Capendale, P.E.; Kersten, V.; Garví, E.S.; Schuurmans, C.C.; Ruelas, M.; Soeiro, J.T.; Tang, G.; Janssen, M.J.; et al. Co-axial printing of convoluted proximal tubule for kidney disease modeling. Biofabrication 2022, 14, 044102. [Google Scholar] [CrossRef]
- He, Q.; Okajima, T.; Onoe, H.; Subagyo, A.; Sueoka, K.; Kuribayashi-Shigetomi, K. Origami-based self-folding of co-cultured NIH/3T3 and HepG2 cells into 3D microstructures. Sci. Rep. 2018, 8, 4556. [Google Scholar] [CrossRef] [Green Version]
- Neal, D.; Sakar, M.S.; Ong, L.-L.S.; Asada, H.H. Formation of elongated fascicle-inspired 3D tissues consisting of high-density, aligned cells using sacrificial outer molding. Lab a Chip 2014, 14, 1907–1916. [Google Scholar] [CrossRef]
- Gori, M.; Giannitelli, S.M.; Torre, M.; Mozetic, P.; Abbruzzese, F.; Trombetta, M.; Traversa, E.; Moroni, L.; Rainer, A. Biofabrication of hepatic constructs by 3D bioprinting of a cell-laden thermogel: An effective tool to assess drug-induced hepatotoxic response. Adv. Health Mater. 2020, 9, 2001163. [Google Scholar] [CrossRef]
- Cui, J.; Wang, H.; Shi, Q.; Sun, T.; Huang, Q.; Fukuda, T. Multicellular co-culture in three-dimensional gelatin methacryloyl hydrogels for liver tissue engineering. Molecules 2019, 24, 1762. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Wang, H.; Zheng, Z.; Shi, Q.; Sun, T.; Huang, Q.; Fukuda, T. Fabrication of perfusable 3D hepatic lobule-like constructs through assembly of multiple cell type laden hydrogel microstructures. Biofabrication 2018, 11, 015016. [Google Scholar] [CrossRef]
- Weng, Y.-S.; Chang, S.-F.; Shih, M.-C.; Tseng, S.-H.; Lai, C.-H. Scaffold-free liver-on-a-chip with multiscale organotypic cultures. Adv. Mater. 2017, 29, 1701545. [Google Scholar] [CrossRef] [PubMed]
- Janani, G.; Mandal, B.B. Mimicking physiologically relevant hepatocyte zonation using immunomodulatory silk liver extracellular matrix scaffolds toward a bioartificial liver platform. ACS Appl. Mater. Interfaces 2021, 13, 24401–24421. [Google Scholar] [CrossRef] [PubMed]
- Fiegel, H.C.; Pryymachuk, G.; Rath, S.; Bleiziffer, O.; Beier, J.P.; Bruns, H.; Kluth, D.; Metzger, R.; Horch, R.E.; Till, H.; et al. Foetal Hepatocyte Transplantation in a Vascularized AV-Loop Transplantation Model in the Rat. J. Cell. Mol. Med. 2010, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khati, V.; Turkki, J.A.; Ramachandraiah, H.; Pati, F.; Gaudenzi, G.; Russom, A. Indirect 3D Bioprinting of a Robust Trilobular Hepatic Construct with Decellularized Liver Matrix Hydrogel. Bioengineering 2022, 9, 603. https://doi.org/10.3390/bioengineering9110603
Khati V, Turkki JA, Ramachandraiah H, Pati F, Gaudenzi G, Russom A. Indirect 3D Bioprinting of a Robust Trilobular Hepatic Construct with Decellularized Liver Matrix Hydrogel. Bioengineering. 2022; 9(11):603. https://doi.org/10.3390/bioengineering9110603
Chicago/Turabian StyleKhati, Vamakshi, Johannes Artturi Turkki, Harisha Ramachandraiah, Falguni Pati, Giulia Gaudenzi, and Aman Russom. 2022. "Indirect 3D Bioprinting of a Robust Trilobular Hepatic Construct with Decellularized Liver Matrix Hydrogel" Bioengineering 9, no. 11: 603. https://doi.org/10.3390/bioengineering9110603