Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Space Flight Information
2.2. Secretome Collection
2.3. Metabolomic Profile of the Secretome
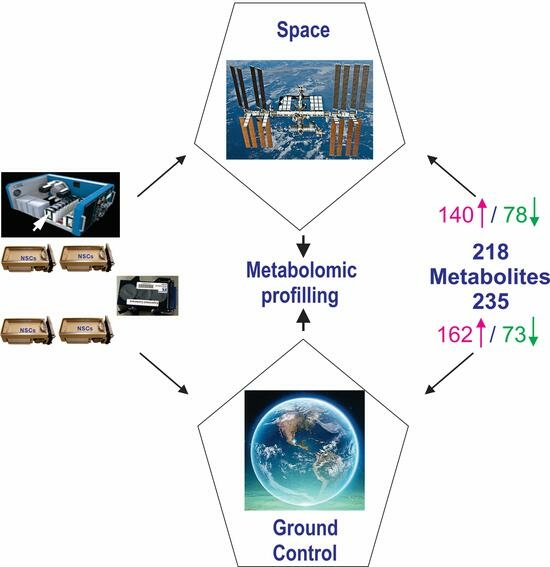
3. Results
3.1. Random Forest and Hierarchical Cluster Analysis
3.2. NSCs in Space (SPC-NSCs) Have High Energy Demands
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaber, E.; Sato, K.; Almeida, E.A. Stem Cell Health and Tissue Regeneration in Microgravity. Stem Cells Dev. 2014, 23, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Finkelstein, H.; Dvorochkin, N.; Sato, K.Y.; Yousuf, R.; Burns, B.P.; Globus, R.K.; Almeida, E.A.; Oss-Ronen, L.; Redden, R.A.; et al. Microgravity Reduces the Differentiation and Regenerative Potential of Embryonic Stem Cells. Stem Cells Dev. 2015, 24, 2605–2621. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.; Blanke, O. The thalamocortical vestibular system in animals and humans. Brain Res. Rev. 2011, 67, 119–146. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, G.G. Cognitive neuroscience in space. Life 2014, 4, 281–294. [Google Scholar] [CrossRef]
- Van Ombergen, A.; Demertzi, A.; Tomilovskaya, E.; Jeurissen, B.; Sijbers, J.; Kozlovskaya, I.B.; Parizel, P.M.; Van de Heyning, P.H.; Sunaert, S.; Laureys, S.; et al. The effect of spaceflight and microgravity on the human brain. J. Neurol. 2017, 264, 18–22. [Google Scholar] [CrossRef]
- Silvano, M.; Miele, E.; Valerio, M.; Casadei, L.; Begalli, F.; Campese, A.F.; Besharat, Z.E.I.N.; Alfano, V.; Abballe, L.; Catanzaro, G.; et al. Consequences of Simulated Microgravity in Neural Stem Cells: Biological Effects and Metabolic Response. J. Stem Cell Res. Ther. 2015, 5, 289. [Google Scholar]
- Cepeda, C.; Vergnes, L.; Carpo, N.; Schibler, M.J.; Bentolila, L.A.; Karouia, F.; Espinosa-Jeffrey, A. Human Neural Stem Cells Flown into Space Proliferate and Generate Young Neurons. Appl. Sci. 2019, 9, 4042. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Lin, H.; Cheng, Y.-C.; Yen, C.-H.; Huang, R.-N.; Lin, K.-H. Beta-adrenoceptor pathway enhances mitochondrial function in human neural stem cells via rotary cell culture system. J. Neurosci. Methods 2012, 207, 130–136. [Google Scholar] [CrossRef]
- Mattei, C.; Alshawaf, A.; D’Abaco, G.; Nayagam, B.; Dottori, M. Generation of Neural Organoids from Human Embryonic Stem Cells Using the Rotary Cell Culture System: Effects of Microgravity on Neural Progenitor Cell Fate. Stem Cells Dev. 2018, 27, 848–857. [Google Scholar] [CrossRef]
- Berardini, M.; Gesualdi, L.; Morabito, C.; Ferranti, F.; Reale, A.; Zampieri, M.; Karpach, K.; Tinari, A.; Bertuccini, L.; Guarnieri, S.; et al. Simulated Microgravity Exposure Induces Antioxidant Barrier Deregulation and Mitochondria Enlargement in TCam-2 Cell Spheroids. Cells 2023, 12, 2106. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Guo, S.; Chai, S.-B.; Yang, J.-Q.; Yue, Y.; Li, H.; Yan, H.-F.; Zhang, T.; Sun, P.-M.; Sun, H.-W.; et al. Proteomic analysis of the effects of simulated microgravity in human gastric mucosal cells. Life Sci. Space Res. 2022, 32, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Jeffrey, A.; Nguyen, K.; Kumar, S.; Toshimasa, O.; Hirose, R.; Reue, K.; Vergnes, L.; Kinchen, J.; de Vellis, J. Simulated microgravity enhances oligodendrocyte mitochondrial function and lipid metabolism. J. Neurosci. Res. 2016, 94, 1434–1450. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Javed, I.; Liu, Y.; Lu, S.; Peng, G.; Zhang, Y.; Qing, H.; Deng, Y. Effect of Prolonged Simulated Microgravity on Metabolic Proteins in Rat Hippocampus: Steps toward Safe Space Travel. J. Proteome Res. 2016, 15, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Jeffrey, A.; Becker-Catania, S.G.; Zhao, P.M.; Cole, R.; Edmond, J.; de Vellis, J. Selective specification of CNS stem cells into oligodendroglial or neuronal cell lineage: Cell culture and transplant studies. J. Neurosci. Res. 2002, 69, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Jeffrey, A.; Wakeman, D.R.; Kim, S.U.; Snyder, E.Y.; de Vellis, J. Culture System for Rodent and Human Oligodendrocyte Specification, Lineage Progression, and Maturation. Curr. Protoc. Stem Cell Biol. 2009, 10, 2D.4.1–2D.4.26. [Google Scholar] [CrossRef] [PubMed]
- Vergnes, L.; Foucaud, B.; Cepeda, C.; Espinosa-Jeffrey, A. Metabolomics Profile of the Secretome of Space-Flown Oligodendrocytes. Cells 2023, 12, 2249. [Google Scholar] [CrossRef]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- DeHaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Chemin. 2010, 2, 1–12. [Google Scholar] [CrossRef]
- Weissgerber, T.L.; Milic, N.M.; Winham, S.J.; Garovic, V.D. Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm. PLoS Biol. 2015, 13, e1002128. [Google Scholar] [CrossRef]
- Mitchell, M.W. Bias of the Random Forest Out-of-Bag (OOB) Error for Certain Input Parameters. Open J. Stat. 2011, 1, 205–211. [Google Scholar] [CrossRef]
- Schurr, A. Lactate: The ultimate cerebral oxidative energy substrate? J. Cereb. Blood Flow Metab. 2006, 26, 142–512. [Google Scholar] [CrossRef] [PubMed]
- Da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Tran, P.H.; Kim, K.-S.; Yang, S.-G. The effects of real and simulated microgravity on cellular mitochondrial function. npj Microgravity 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Costantini, D.; Overi, D.; Casadei, L.; Cardinale, V.; Nevi, L.; Carpino, G.; Di Matteo, S.; Safarikia, S.; Valerio, M.; Melandro, F.; et al. Simulated microgravity promotes the formation of tridimensional cultures and stimulates pluripotency and a glycolytic metabolism in human hepatic and biliary tree stem/progenitor cells. Sci. Rep. 2019, 9, 5559. [Google Scholar] [CrossRef] [PubMed]
- Krebs, H.A.; Weitzman, P.D.J. Krebs’ Citric Acid Cycle: Half a Century and Still Turning; Biochemical Society: London, UK, 1987; p. 25. [Google Scholar]
- Yang, Y.; Sauve, A.A. NAD + metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2016, 1864, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Bardócz, S.; Duguid, T.J.; Brown, D.S.; Grant, G.; Pusztai, A.; White, A.; Ralph, A. The importance of dietary polyamines in cell regeneration and growth. Br. J. Nutr. 1995, 73, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010, 42, 39–51. [Google Scholar] [CrossRef]
- Pegg, A.E. Functions of Polyamines in Mammals. J. Biol. Chem. 2016, 291, 14904–14912. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Pietrocola, F.; Lachkar, S.; Enot, D.P.; Niso-Santano, M.; Pedro, J.M.B.-S.; Sica, V.; Izzo, V.; Maiuri, M.C.; Madeo, F.; Mariño, G.; et al. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 2015, 22, 509–516. [Google Scholar] [CrossRef]
- Yatagai, F.; Honma, M.; Dohmae, N.; Ishioka, N. Biological effects of space environmental factors: A possible interaction between space radiation and microgravity. Life Sci. Space Res. 2019, 20, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, A.; McDonald, J.T.; Hada, M.; Takahashi, A.; Mason, C.E.; Mognato, M. Genomic Changes Driven by Radiation-Induced DNA Damage and Microgravity in Human Cells. Int. J. Mol. Sci. 2021, 22, 10507. [Google Scholar] [CrossRef] [PubMed]
- Laiakis, E.C.; Shuryak, I.; Deziel, A.; Wang, Y.-W.; Barnette, B.L.; Yu, Y.; Ullrich, R.L.; Fornace, A.J.; Emmett, M.R. Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome. Int. J. Mol. Sci. 2021, 22, 3070. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.W.; Nishiyama, N.C.; Pecaut, M.J.; Campbell-Beachler, M.; Gifford, P.; Haynes, K.E.; Becronis, C.; Gridley, D.S. Simulated Microgravity and Low-Dose/Low-Dose-Rate Radiation Induces Oxidative Damage in the Mouse Brain. Radiat. Res. 2016, 185, 647–657. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biancotti, J.C.; Espinosa-Jeffrey, A. Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space. Bioengineering 2024, 11, 11. https://doi.org/10.3390/bioengineering11010011
Biancotti JC, Espinosa-Jeffrey A. Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space. Bioengineering. 2024; 11(1):11. https://doi.org/10.3390/bioengineering11010011
Chicago/Turabian StyleBiancotti, Juan Carlos, and Araceli Espinosa-Jeffrey. 2024. "Metabolomic Profiling of the Secretome from Human Neural Stem Cells Flown into Space" Bioengineering 11, no. 1: 11. https://doi.org/10.3390/bioengineering11010011