Xanthine Oxidase Inhibitory Peptides from Larimichthys polyactis: Characterization and In Vitro/In Silico Evidence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Pretreatment of Raw Materials
2.3. Determination of Raw Materials Protein
2.4. Preparation of Papain Hydrolysates from SYC
2.5. Preparation of Peptide Fractions of SYCHs
2.6. Determination of Amino Acids Composition of SYC and SYCHs
2.7. Determination of XOI Activity IC50 In Vitro
2.8. Determination of MW Distributions
2.9. Identification of the AA Sequence and Molecular Mass of SYCH
2.10. Physicochemical Properties Prediction of Active Peptides
2.11. Peptides Synthesis
2.12. Molecular Docking and Interaction Visual Analysis
2.13. Statistical Analysis
3. Results and Discussion
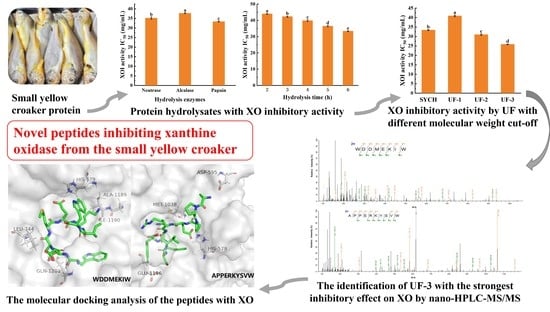
3.1. The Potential of SYC to Decrease Uric Acid Levels
3.2. The Optimal Conditions for the Preparation of SYCH
3.3. MW Distribution of XOI Peptides
3.4. Identification of XOI Peptides Sequences and Validation of XOI Activity
3.5. Molecular Docking and Visual Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaneko, K.; Takayanagi, F.; Fukuuchi, T.; Yamaoka, N.; Yasuda, M.; Mawatari, K.I.; Fujimori, S. Determination of total purine and purine base content of 80 food products to aid nutritional therapy for gout and hyperuricemia. Nucleosides Nucleotides Nucleic Acids 2020, 39, 1449–1457. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Yuan, M.; Xu, Y.; Xu, H. Gout and diet: A comprehensive review of mechanisms and management. Nutrients 2022, 14, 3525. [Google Scholar] [CrossRef]
- Lee, Y.; Hwang, J.; Desai, S.H.; Li, X.; Jenkins, C.; Kopp, J.B.; Winkler, C.A.; Cho, S.K. Efficacy of xanthine oxidase inhibitors in lowering serum uric acid in chronic kidney disease: A systematic review and meta-analysis. J. Clin. Med. 2022, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Su, G.; Sun-Waterhouse, D.; Waterhouse, G.I.N.; Zhao, M.; Liu, Y. In vivo anti-hyperuricemic and xanthine oxidase inhibitory properties of tuna protein hydrolysates and its isolated fractions. Food Chem. 2019, 272, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Ichida, K.; Matsuo, H.; Takada, T.; Nakayama, A.; Murakami, K.; Shimizu, T.; Yamanashi, Y.; Kasuga, H.; Nakashima, H.; Nakamura, T.; et al. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat. Commun. 2012, 3, 764. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Zhu, D.; Sun-Waterhouse, D.; Su, G.; Lin, L.; Wang, X.; Dong, Y. In vitro and in vivo studies on adlay-derived seed extracts: Phenolic profiles, antioxidant activities, serum uric acid suppression, and xanthine oxidase inhibitory effects. J. Agric. Food Chem. 2014, 62, 7771–7778. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Y.; Zhong, H.; Chen, F.; Regenstein, J.; Hu, X.; Cai, L.; Feng, F. The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategies. Crit. Rev. Food Sci. Nutr. 2022, 62, 3979–3989. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Wang, F.; Xing, C. A zebrafish (danio rerio) model for high-throughput screening food and drugs with uric acid-lowering activity. Biochem. Biophys. Res. Commun. 2019, 508, 494–498. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Fitzgerald, R.J. Tryptophan-containing milk protein-derived dipeptides inhibit xanthine oxidase. Peptides 2012, 37, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Shi, C.; Wang, M.; Zhou, M.; Liang, M.; Zhang, T.; Yuan, E.; Wang, Z.; Yao, M.; Ren, J. Tryptophan residue enhances in vitro walnut protein-derived peptides exerting xanthine oxidase inhibition and antioxidant activities. J. Funct. Foods 2019, 53, 276–285. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, Y.; Liu, J.; Hu, Z.; Du, Y.; Sun, J.; Mao, X. Separation, identification and docking analysis of xanthine oxidase inhibitory peptides from pacific cod bone-flesh mixture. LWT 2022, 167, 113862. [Google Scholar] [CrossRef]
- Zhao, Q.; Jiang, X.; Mao, Z.; Zhang, J.; Sun, J.; Mao, X. Exploration, sequence optimization and mechanism analysis of novel xanthine oxidase inhibitory peptide from Ostrea rivularis Gould. Food Chem. 2022, 404, 134537. [Google Scholar] [CrossRef]
- Zhong, H.; Abdullah; Zhang, Y.; Deng, L.; Zhao, M.; Tang, J.; Zhang, H.; Feng, F.; Wang, J. Exploring the potential of novel xanthine oxidase inhibitory peptide (ACECD) derived from Skipjack tuna hydrolysates using affinity-ultrafiltration coupled with HPLC-MALDI-TOF/TOF-MS. Food Chem. 2021, 347, 129068. [Google Scholar] [CrossRef]
- Hu, X.; Zhou, Y.; Zhou, S.; Chen, S.; Wu, Y.; Li, L.; Yang, X. Purification and identification of novel xanthine oxidase inhibitory peptides derived from Round Scad (Decapterus maruadsi) protein hydrolysates. Mar. Drugs 2021, 19, 538. [Google Scholar] [CrossRef]
- Jia, L.; Wang, L.; Liu, C.; Liang, Y.; Lin, Q. Bioactive peptides from foods: Production, function, and application. Food Funct. 2021, 12, 7108–7125. [Google Scholar] [CrossRef]
- Cunha, S.A.; Pintado, M.E. Bioactive peptides derived from marine sources: Biological and functional properties. Trends Food Sci. Technol. 2022, 119, 348–370. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Tayyab Rashid, M.; Yan, J.K.; Ma, H. Effect of multi-frequency ultrasound thawing on the structure and rheological properties of myofibrillar proteins from small yellow croaker. Ultrason. Sonochem. 2021, 70, 105352. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zeng, W.; Rong, Y.; Lou, B. Compositions, nutritional and texture quality of wild-caught andcage-cultured small yellow croaker. J. Food Compos. Anal. 2022, 107, 104370. [Google Scholar] [CrossRef]
- Allegrini, S.; Garcia-Gil, M.; Pesi, R.; Camici, M.; Tozzi, M.G. The good, the bad and the new about uric acid in cancer. Cancers 2022, 14, 4959. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Oh, B.J.; Lee, J.H.; Yoon, Y.S.; Lee, H.I. Effects of various drying methods on physicochemical characteristics and textural features of yellow croaker (Larimichthys polyactis). Foods 2020, 9, 196. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.-M.; Li, J.; Zhao, Q.; Lv, D.-D.; Rakariyatham, K. The effect of frying process on lipids in small yellow croaker (Larimichthys polyactis) and frying oil. J. Aquat. Food Prod. Technol. 2021, 31, 83–95. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, M.; Chen, G.; Wang, Y. Effect of micronization on physicochemical properties of small yellow croaker (Pseudosciaena polyactis) skull. Adv. Powder. Technol. 2013, 24, 932–938. [Google Scholar] [CrossRef]
- Hou, M.; Xiang, H.; Hu, X.; Chen, S.; Wu, Y.; Xu, J.; Yang, X. Novel potential XOD inhibitory peptides derived from Trachinotus ovatus: Isolation, identification and structure-function analysis. Food Biosci. 2022, 47, 101639. [Google Scholar] [CrossRef]
- Liu, N.; Wang, Y.; Yang, M.; Bian, W.; Zeng, L.; Yin, S.; Xiong, Z.; Hu, Y.; Wang, S.; Meng, B.; et al. New rice-derived short peptide potently alleviated hyperuricemia induced by potassium oxonate in rats. J. Agric. Food Chem. 2019, 67, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ji, H.; Song, W.; Peng, S.; Zhan, S.; Qu, Y.; Chen, M.; Zhang, D.; Liu, S. Hypouricemic, hepatoprotective and nephroprotective roles of oligopeptides derived from Auxis thazard protein in hyperuricemic mice. Food Funct. 2021, 12, 11838–11848. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Si, X.; Ding, X.; Duan, L.; Xiao, C. pH-responsive hydrogels based on the self-assembly of short polypeptides for controlled release of peptide and protein drugs. J. Polym. Res. 2019, 26, 278. [Google Scholar] [CrossRef]
- Çağlar, A.F.; Çakır, B.; Gülseren, İ. LC-Q-TOF/MS based identification and in silico verification of ACE-inhibitory peptides in Giresun (Turkey) hazelnut cakes. Eur. Food Res. Technol. 2021, 247, 1189–1198. [Google Scholar] [CrossRef]
- Yu, Z.; Kan, R.; Wu, S.; Guo, H.; Zhao, W.; Ding, L.; Zheng, F.; Liu, J. Xanthine oxidase inhibitory peptides derived from tuna protein: Virtual screening, inhibitory activity, and molecular mechanisms. J. Sci. Food Agr. 2021, 101, 1349–1354. [Google Scholar] [CrossRef]
- Gui, M.; Gao, L.; Rao, L.; Li, P.; Zhang, Y.; Han, J.W.; Li, J. Bioactive peptides identified from enzymatic hydrolysates of sturgeon skin. J. Sci. Food Agr. 2022, 102, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Fan, S.; Lu, G.; Sun, N.; Wang, R.; Lu, C.; Han, J.; Zhou, J.; Li, Y.; Ming, T.; et al. Systematic investigation of the amino acid profiles that are correlated with xanthine oxidase inhibitory activity: Effects, mechanism and applications in protein source screening. Free Radic. Biol. Med. 2021, 177, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, X.; Shi, C.; Li, Y.; Majumder, K.; Ning, Z.; Ren, J. Moderation of hyperuricemia in rats via consuming walnut protein hydrolysate diet and identification of new antihyperuricemic peptides. Food Funct. 2018, 9, 107–116. [Google Scholar] [CrossRef]
- Allahyari, M.; Samadi-Noshahr, Z.; Hosseinian, S.; Salmani, H.; Noras, M.; Khajavi-Rad, A. Camel milk and allopurinol attenuated adenine-induced acute renal failure in rats. Iran. J. Sci. Technol. A 2021, 45, 1539–1548. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Wang, J.; Liu, H.; Kwong, J.S.; Chen, H.; Li, L.; Chung, S.C.; Shah, A.; Chen, Y.; et al. Diagnosis and treatment for hyperuricemia and gout: A systematic review of clinical practice guidelines and consensus statements. BMJ Open 2019, 9, e026677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalamaiah, M.; Dinesh Kumar, B.; Hemalatha, R.; Jyothirmayi, T. Fish protein hydrolysates: Proximate composition, amino acid composition, antioxidant activities and applications: A review. Food Chem. 2012, 135, 3020–3038. [Google Scholar] [CrossRef]
- Elavarasan, K.; Shamasundar, B.A. Antioxidant properties of papain mediated protein hydrolysates from fresh water carps (Catla catla, Labeo rohita and Cirrhinus mrigala) and its application on inhibition of lipid oxidation in oil sardine mince during ice storage. J. Food Sci. Technol. 2022, 59, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kang, X.; Li, Q.; Shi, C.; Lian, Y.; Yuan, E.; Zhou, M.; Ren, J. Anti-hyperuricemic peptides derived from bonito hydrolysates based on in vivo hyperuricemic model and in vitro xanthine oxidase inhibitory activity. Peptides 2018, 107, 45–53. [Google Scholar] [CrossRef]
- Wei, L.; Ji, H.; Song, W.; Peng, S.; Zhan, S.; Qu, Y.; Chen, M.; Zhang, D.; Liu, S. Identification and molecular docking of two novel peptides with xanthine oxidase inhibitory activity from Auxis thazard. Food Sci. Technol. 2022, 42, e106921. [Google Scholar] [CrossRef]
- Vásquez, P.; Zapata, J.E.; Chamorro, V.C.; García Fillería, S.F.; Tironi, V.A. Antioxidant and angiotensin I-converting enzyme (ACE) inhibitory peptides of rainbow trout (Oncorhynchus mykiss) viscera hydrolysates subjected to simulated gastrointestinal digestion and intestinal absorption. LWT 2022, 154, 112834. [Google Scholar] [CrossRef]
- Etemadian, Y.; Ghaemi, V.; Shaviklo, A.R.; Pourashouri, P.; Sadeghi Mahoonak, A.R.; Rafipour, F. Development of animal/ plant-based protein hydrolysate and its application in food, feed and nutraceutical industries: State of the art. J. Clean. Prod. 2021, 278, 123219. [Google Scholar] [CrossRef]
- Islam, M.S.; Wang, H.; Admassu, H.; Sulieman, A.A.; Wei, F.A. Health benefits of bioactive peptides produced from muscle proteins: Antioxidant, anti-cancer, and anti-diabetic activities. Process Biochem. 2022, 116, 116–125. [Google Scholar] [CrossRef]
- Wu, Y.; He, H.; Hou, T. Purification, identification, and computational analysis of xanthine oxidase inhibitory peptides from kidney bean. J. Food Sci. 2021, 86, 1081–1088. [Google Scholar] [CrossRef] [PubMed]





| AA | Content (mg/g) | Proportion | AA | Content (mg/g) | Proportion |
|---|---|---|---|---|---|
| Glu | 173.82 ± 1.00 | 21.08% | Val | 37.03 ± 0.25 | 4.49% |
| Asp | 77.60 ± 0.66 | 9.41% | Phe | 35.60 ± 0.83 | 4.32% |
| Lys | 74.77 ± 0.51 | 9.07% | Ser | 35.68 ± 0.25 | 4.33% |
| Leu | 66.83 ± 0.75 | 8.11% | Tyr | 31.02 ± 0.47 | 3.76% |
| Ala | 52.98 ± 0.43 | 6.43% | Met | 28.06 ± 0.55 | 3.40% |
| Arg | 52.85 ± 0.53 | 6.41% | His | 15.65 ± 0.25 | 1.90% |
| Thr | 37.78 ± 0.99 | 4.58% | Pro | 14.31 ± 0.43 | 1.74% |
| Gly | 38.05 ± 0.75 | 4.62% | Cys | 5.08 ± 0.13 | 0.62% |
| Ile | 36.53 ± 0.87 | 4.43% | Trp | 10.78 ± 0.86 | 1.31% |
| EAA | 327.39 ± 5.41 | 39.71% | HAA | 282.14 ± 4.85 | 34.22% |
| BAA | 143.27 ± 1.24 | 17.38% | AAA | 77.40 ± 2.10 | 9.39% |
| TAA | 824.45 ± 10.50 | 100% |
| Samples | SYCH | UF-3 | Samples | SYCH | UF-3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| AA | Content (mg/g) | Proportion | Content (mg/g) | Proportion | AA | Content (mg/g) | Proportion | Content (mg/g) | Proportion |
| Glu | 164.78 ± 1.77 | 21.05% | 127.27 ± 3.02 | 18.87% | Ile | 26.50 ± 1.23 | 3.39% | 24.01 ± 0.40 | 3.56% |
| Lys | 79.46 ± 1.57 | 10.15% | 68.56 ± 0.65 | 10.17% | Tyr | 24.80 ± 0.31 | 3.17% | 22.98 ± 0.24 | 3.41% |
| Asp | 80.55 ± 2.18 | 10.29% | 63.64 ± 0.89 | 9.44% | His | 17.77 ± 0.97 | 2.27% | 17.25 ± 0.32 | 2.56% |
| Leu | 63.08 ± 0.89 | 8.06% | 54.77 ± 0.16 | 8.12% | Met | 17.63 ± 1.03 | 2.25% | 14.66 ± 0.60 | 2.17% |
| Ala | 57.23 ± 1.03 | 7.31% | 55.45 ± 1.26 | 8.22% | Pro | 19.77 ± 0.91 | 2.53% | 15.92 ± 0.34 | 2.36% |
| Arg | 50.84 ± 0.76 | 6.49% | 45.18 ± 0.40 | 6.70% | Cys | 5.28 ± 0.53 | 0.67% | 5.45 ± 0.28 | 0.81% |
| Thr | 37.12 ± 1.20 | 4.74% | 31.67 ± 0.17 | 4.70% | EAA | 288.50 ± 8.57 | 36.86% | 255.81 ± 1.13 | 37.84% |
| Gly | 36.86 ± 0.30 | 4.71% | 33.86 ± 0.94 | 5.02% | HAA | 248.92 ± 6.66 | 31.80% | 228.32 ± 2.24 | 33.86% |
| Ser | 36.36 ± 0.68 | 4.64% | 30.17 ± 0.11 | 4.47% | AAA | 54.67 ± 1.67 | 6.99% | 49.81 ± 0.02 | 7.39% |
| Val | 34.83 ± 1.45 | 4.45% | 36.67 ± 0.99 | 5.44% | BAA | 148.07 ± 3.00 | 18.92% | 130.98 ± 1.32 | 19.42% |
| Phe | 29.88 ± 1.41 | 3.82% | 26.83 ± 0.23 | 3.98% | TAA | 782.73 ± 16.20 | 100% | 676.00 ± 2.96 | 100% |
| Serial Number | Amino Acid Sequence | Amino Acid Number | Molecular Weight | Isoelectric Point | Predicted Activity Values | Hemolysis | Toxicity | XOI Activity of IC50 (mM) |
|---|---|---|---|---|---|---|---|---|
| 1 | WDDMEKIW | 8 | 1122.25 | 4.03 | 0.73 | Non-hemolytic | Non-toxic | 3.16 ± 0.03 |
| 2 | APPERKYSVW | 10 | 1232.40 | 8.63 | 0.58 | Non-hemolytic | Non-toxic | 5.86 ± 0.02 |
| 3 | IADRMQKELT | 10 | 1204.41 | 6.07 | 0.15 | Non-hemolytic | Non-toxic | NA |
| 4 | LNSADLIK | 8 | 873.02 | 5.84 | 0.19 | Non-hemolytic | Non-toxic | NA |
| 5 | LSNLGIVI | 8 | 828.02 | 5.52 | 0.18 | Hemolytic | Non-toxic | NA |
| 6 | IGALRAVA | 8 | 769.94 | 9.75 | 0.14 | Hemolytic | Non-toxic | NA |
| 7 | HHTFYNELR | 9 | 1216.3 | 6.92 | 0.38 | Non-hemolytic | Non-toxic | NA |
| Peptides | Binding Energy (kcal/mol) | Hydrogen Bonding | Hydrophobic Interactions | Electrostatic Interaction |
|---|---|---|---|---|
| WDDMEKIW | −7.3 | Ile1190, Ala1189, Leu744, Gln1201 | Val1200, Gly1197, Glu1196, Phe1219, Ile1235, Ile1229, Phe1232, Pro1230, His741, Ala1231, Tyr743, Phe238, Phe742, Tyr592, Met1038, Gly1039, Gly796, Gly1039, Met794, Gly795, Ala582, Gln585, Gln1194, Gly1193 | His579 |
| APPERKYSVW | −7.9 | Arg912, Met1038 | Ala582, His579, Gln585, Met794, Gly796, Gly795, Leu744, Tyr743, Tyr592, Gly039, Gln194, Gly193, Gln021, Phe798, Ala1198, Glu1196, Ile1235, Phe1239, Gly1197, Val1200, Phe1232, Ala1231 | NP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Guan, W.; Li, Y.; Zhang, J.; Cai, L. Xanthine Oxidase Inhibitory Peptides from Larimichthys polyactis: Characterization and In Vitro/In Silico Evidence. Foods 2023, 12, 982. https://doi.org/10.3390/foods12050982
Chen X, Guan W, Li Y, Zhang J, Cai L. Xanthine Oxidase Inhibitory Peptides from Larimichthys polyactis: Characterization and In Vitro/In Silico Evidence. Foods. 2023; 12(5):982. https://doi.org/10.3390/foods12050982
Chicago/Turabian StyleChen, Xiaoling, Weiliang Guan, Yujin Li, Jinjie Zhang, and Luyun Cai. 2023. "Xanthine Oxidase Inhibitory Peptides from Larimichthys polyactis: Characterization and In Vitro/In Silico Evidence" Foods 12, no. 5: 982. https://doi.org/10.3390/foods12050982