A Rapid Non-Destructive Hyperspectral Imaging Data Model for the Prediction of Pungent Constituents in Dried Ginger
Abstract
:1. Introduction
2. Data and Methods
2.1. Sample Collection, Preparation, and Chemical Analysis
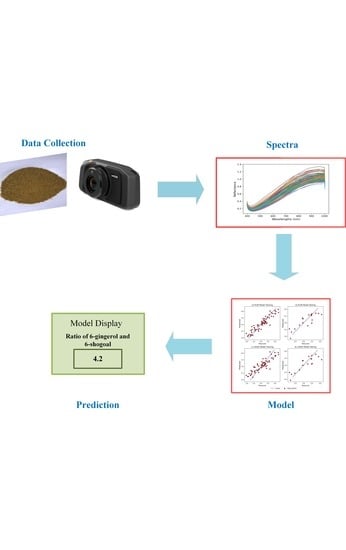
2.2. Hyperspectral Imaging System and Acquisition
2.3. Image Correction and Spectrum Extraction
2.4. Spectral Pre-Processing
2.5. Predictive Modeling and Validation
2.5.1. Partial Least Squares Regression
2.5.2. LASSO Regression
2.5.3. Model Evaluation
2.6. Wavelength Selection Method
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddaraju, M.N.; Dharmesh, S.M. Inhibition of gastric H+,K+-ATPase andHelicobacter pylori growth by phenolic antioxidants of Zingiber officinale. Mol. Nutr. Food Res. 2007, 51, 324–332. [Google Scholar] [CrossRef]
- Grzanna, R.; Lindmark, L.; Frondoza, C.G. Ginger—An Herbal Medicinal Product with Broad Anti-Inflammatory Actions. J. Med. Food 2005, 8, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ghayur, M.N.; Gilani, A. Ginger Lowers Blood Pressure Through Blockade of Voltage-Dependent Calcium Channels. J. Cardiovasc. Pharmacol. 2005, 45, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Nurtjahja-Tjendraputra, E.; Ammit, A.; Roufogalis, B.; Tran, V.H.; Duke, C.C. Effective anti-platelet and COX-1 enzyme inhibitors from pungent constituents of ginger. Thromb. Res. 2003, 111, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Ojewole, J.A. Analgesic, anti-inflammatory and hypoglycaemic effects of ethanol extract of Zingiber officinale (Roscoe) rhizomes (Zingiberaceae) in mice and rats. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2006, 20, 764–772. [Google Scholar]
- Nair, K.P. Turmeric (Curcuma Longa L.) and Ginger (Zingiber officinale Rosc.)–World’s Invaluable Medicinal Spices: The Agronomy and Economy of Turmeric and Ginger; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Kumara, M.; Shylajab, M.; Nazeemc, P.; Babu, T. 6-Gingerol is the most potent anticancerous compound in ginger (Zingiber officinale Rosc.). J. Dev. Drugs 2017, 6, 1000167. [Google Scholar]
- Jiang, Y.; Huang, M.; Wisniewski, M.; Li, H.; Zhang, M.; Tao, X.; Liu, Y.; Zou, Y. Transcriptome Analysis Provides Insights into Gingerol Biosynthesis in Ginger (Zingiber officinale). Plant Genome 2018, 11, 180034. [Google Scholar] [CrossRef] [Green Version]
- Yudthavorasit, S.; Wongravee, K.; Leepipatpiboon, N. Characteristic fingerprint based on gingerol derivative analysis for discrimination of ginger (Zingiber officinale) according to geographical origin using HPLC-DAD combined with chemometrics. Food Chem. 2014, 158, 101–111. [Google Scholar] [CrossRef]
- Johnson, J.B.; Mani, J.S.; Naiker, M. Gingerol, Shogaol and Paradol: The Chemistry of Pungent Ginger Constituents. In Proceedings of the Queensland Annual Chemistry Symposium, Queensland, Australia, 27 November 2020; pp. 89–90. [Google Scholar]
- Narasimhan, S.; Govindarajan, V.S. Evaluation of spices and oleoresin-VI-pungency of ginger components, gingerols and shogoals and quality. Int. J. Food Sci. Technol. 2007, 13, 31–36. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Jaafar, H.Z.; Baghdadi, A.; Tayebi-Meigooni, A. Formation of 6-, 8- and 10-Shogaol in Ginger through Application of Different Drying Methods: Altered Antioxidant and Antimicrobial Activity. Molecules 2018, 23, 1646. [Google Scholar] [CrossRef] [Green Version]
- Sanwal, S.; Rai, N.; Singh, J.; Buragohain, J. Antioxidant phytochemicals and gingerol content in diploid and tetraploid clones of ginger (Zingiber officinale Roscoe). Sci. Hortic. 2010, 124, 280–285. [Google Scholar] [CrossRef]
- Wohlmuth, H.; Leach, D.N.; Smith, M.K.; Myers, S.P. Gingerol Content of Diploid and Tetraploid Clones of Ginger (Zingiber officinale Roscoe). J. Agric. Food Chem. 2005, 53, 5772–5778. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-C.; Chung, C.-C.; Wang, H.-Y.; Law, C.-L.; Chen, H.-H. Formation of 6-Shogaol of Ginger Oil Under Different Drying Conditions. Dry. Technol. 2011, 29, 1884–1889. [Google Scholar] [CrossRef]
- Pawar, N.V.; Pai, S.R.; Nimbalkar, M.S.; Dixit, G.B. RP-HPLC analysis of phenolic antioxidant compound 6-gingerol from in vitro cultures of Zingiber officinale Roscoe. Plant Sci. Today 2015, 2, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Khoo, C.; Halstead, C.W.; Huynh, T.; Bensoussan, A. Liquid chromatographic determination of 6-, 8-, 10-gingerol, and 6-shogaol in ginger (Zingiber officinale) as the raw herb and dried aqueous extract. J. AOAC Int. 2007, 90, 1219–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cafino, E.J.V.; Lirazan, M.B.; Marfori, E.C. A simple HPLC method for the analysis of [6]-gingerol produced by multiple shoot culture of ginger (Zingiber officinale). Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 38–42. [Google Scholar]
- Su, W.-H.; Sun, D.-W. Fourier Transform Infrared and Raman and Hyperspectral Imaging Techniques for Quality Determinations of Powdery Foods: A Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 104–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pu, H.; Sun, D.-W. Hyperspectral imaging technique for evaluating food quality and safety during various processes: A review of recent applications. Trends Food Sci. Technol. 2017, 69, 25–35. [Google Scholar] [CrossRef]
- Caporaso, N.; ElMasry, G.; Gou, P. Hyperspectral imaging techniques for noncontact sensing of food quality. In Innovative Food Analysis; Elsevier BV: Amsterdam, The Netherlands, 2021; pp. 345–379. [Google Scholar]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [Green Version]
- Su, W.-H.; Bakalis, S.; Sun, D.-W. Chemometrics in tandem with near infrared (NIR) hyperspectral imaging and Fourier transform mid infrared (FT-MIR) microspectroscopy for variety identification and cooking loss determination of sweet potato. Biosyst. Eng. 2019, 180, 70–86. [Google Scholar] [CrossRef]
- Su, W.-H.; Xue, H. Imaging Spectroscopy and Machine Learning for Intelligent Determination of Potato and Sweet Potato Quality. Foods 2021, 10, 2146. [Google Scholar] [CrossRef] [PubMed]
- Su, W.-H.; Yang, C.; Dong, Y.; Johnson, R.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Hyperspectral imaging and improved feature variable selection for automated determination of deoxynivalenol in various genetic lines of barley kernels for resistance screening. Food Chem. 2020, 343, 128507. [Google Scholar] [CrossRef]
- Rodríguez-Pulido, F.J.; Hernández-Hierro, J.M.; Nogales-Bueno, J.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. A novel method for evaluating flavanols in grape seeds by near infrared hyperspectral imaging. Talanta 2014, 122, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Ariana, D.P.; Lu, R.; Guyer, D.E. Near-infrared hyperspectral reflectance imaging for detection of bruises on pickling cucumbers. Comput. Electron. Agric. 2006, 53, 60–70. [Google Scholar] [CrossRef]
- Su, W.-H.; Bakalis, S.; Sun, D.-W. Potato hierarchical clustering and doneness degree determination by near-infrared (NIR) and attenuated total reflectance mid-infrared (ATR-MIR) spectroscopy. J. Food Meas. Charact. 2019, 13, 1218–1231. [Google Scholar] [CrossRef]
- Mahesh, S.; Jayas, D.; Paliwal, J.; White, N. Hyperspectral imaging to classify and monitor quality of agricultural materials. J. Stored Prod. Res. 2015, 61, 17–26. [Google Scholar] [CrossRef]
- Pu, Y.-Y.; Sun, D.-W. Vis–NIR hyperspectral imaging in visualizing moisture distribution of mango slices during microwave-vacuum drying. Food Chem. 2015, 188, 271–278. [Google Scholar] [CrossRef]
- Pu, Y.-Y.; Zhao, M.; O’Donnell, C.; Sun, D.-W. Non-destructive quality evaluation of banana slices during microwave vacuum drying using spectral and imaging techniques. Dry. Technol. 2018, 36, 1542–1553. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Fan, Y.; Tao, X.; Yin, W.; He, Y. Hyperspectral Imaging for Predicting the Internal Quality of Kiwifruits Based on Variable Selection Algorithms and Chemometric Models. Sci. Rep. 2017, 7, 7845. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Pulido, F.J.; Gordillo, B.; Heredia, F.J.; González-Miret, M.L. CIELAB—Spectral image MATCHING: An app for merging colorimetric and spectral images for grapes and derivatives. Food Control 2021, 125, 108038. [Google Scholar] [CrossRef]
- Lin, X.; Sun, D.-W. Investigation of moisture distribution of ginger slices and splits during hot-air drying and rehydration procedures by NIR hyperspectral imaging. In Sensing for Agriculture and Food Quality and Safety XII; SPIE: Bellingham, WA, USA, 2020; Volume 11421, p. 114210D. [Google Scholar]
- Lin, X.; Xu, J.-L.; Sun, D.-W. Comparison of moisture uniformity between microwave-vacuum and hot-air dried ginger slices using hyperspectral information combined with semivariogram. Dry. Technol. 2021, 39, 1044–1058. [Google Scholar] [CrossRef]
- Lin, X.; Xu, J.-L.; Sun, D.-W. Evaluating drying feature differences between ginger slices and splits during microwave-vacuum drying by hyperspectral imaging technique. Food Chem. 2020, 332, 127407. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, P.-H.; Zhou, G.-S.; Wang, Y.-J.; Bao, B.-H.; Wu, Q.-N.; Huang, S.-L. Rapid and practical qualitative and quantitative evaluation of non-fumigated ginger and sulfur-fumigated ginger via Fourier-transform infrared spectroscopy and chemometric methods. Food Chem. 2021, 341, 128241. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.B.; Mani, J.S.; White, S.; Brown, P.; Naiker, M. Quantitative profiling of gingerol and its derivatives in Australian ginger. J. Food Compos. Anal. 2021, 104, 104190. [Google Scholar] [CrossRef]
- Johnson, J.B.; Mani, J.S.; White, S.; Brown, P.; Naiker, M. Pungent and volatile constituents of dried Australian ginger. Curr. Res. Food Sci. 2021, 4, 612–618. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, F.; He, Y. Identification of coffee bean varieties using hyperspectral imaging: Influence of pre-processing methods and pixel-wise spectra analysis. Sci. Rep. 2018, 8, 2166. [Google Scholar] [CrossRef]
- Buddenbaum, H.; Steffens, M. The effects of spectral pre-treatments on chemometric analyses of soil profiles using laboratory imaging spectroscopy. Appl. Environ. Soil Sci. 2012, 2012, 274903. [Google Scholar] [CrossRef] [Green Version]
- Jung, A.; Vohland, M.; Thiele-Bruhn, S. Use of A Portable Camera for Proximal Soil Sensing with Hyperspectral Image Data. Remote Sens. 2015, 7, 11434–11448. [Google Scholar] [CrossRef] [Green Version]
- Kämper, W.; Trueman, S.J.; Tahmasbian, I.; Bai, S.H. Rapid Determination of Nutrient Concentrations in Hass Avocado Fruit by Vis/NIR Hyperspectral Imaging of Flesh or Skin. Remote Sens. 2020, 12, 3409. [Google Scholar] [CrossRef]
- Malmir, M.; Tahmasbian, I.; Xu, Z.; Farrar, M.B.; Bai, S.H. Prediction of soil macro- and micro-elements in sieved and ground air-dried soils using laboratory-based hyperspectral imaging technique. Geoderma 2019, 340, 70–80. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Walvoort, D.J.J.; McBratney, A.B.; Janik, L.J.; Skjemstad, J.O. Visible, near infrared, mid infrared or combined diffuse reflectance spectroscopy for simultaneous assessment of various soil properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Vasques, G.; Grunwald, S.; Sickman, J. Comparison of multivariate methods for inferential modeling of soil carbon using visible/near-infrared spectra. Geoderma 2008, 146, 14–25. [Google Scholar] [CrossRef]
- Sädekoski, N. Tropical Altitudinal Gradient Soil Organic Carbon Estimation with VIS-NIR (400–1000 nm) Imaging Spectroscopy; Helsingin Yliopisto: Helsinki, Finland, 2020. [Google Scholar]
- Lazaridis, D.C.; Verbesselt, J.; Robinson, A.P. Penalized regression techniques for prediction: A case study for predicting tree mortality using remotely sensed vegetation indices. Can. J. For. Res. 2011, 41, 24–34. [Google Scholar] [CrossRef]
- Cheng, J.-H.; Sun, D.-W. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle. Food Eng. Rev. 2017, 9, 36–49. [Google Scholar] [CrossRef]
- Lius, Y. Prediction of nitrogen and phosphorus contents in citrus leaves based on hyperspectral imaging. Int. J. Agric. Biol. Eng. 2015, 8, 80–88. [Google Scholar]
- Malmir, M.; Tahmasbian, I.; Xu, Z.; Farrar, M.B.; Bai, S.H. Prediction of macronutrients in plant leaves using chemometric analysis and wavelength selection. J. Soils Sediments 2020, 20, 249–259. [Google Scholar] [CrossRef]
- Abdi, H. Partial Least Squares (PLS) Regression. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 97–106. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Bellon-Maurel, V.; Fernandez-Ahumada, E.; Palagos, B.; Roger, J.-M.; McBratney, A. Critical review of chemometric indicators commonly used for assessing the quality of the prediction of soil attributes by NIR spectroscopy. TrAC Trends Anal. Chem. 2010, 29, 1073–1081. [Google Scholar] [CrossRef]
- Iqbal, A.; Sun, D.-W.; Allen, P. Prediction of moisture, color and pH in cooked, pre-sliced turkey hams by NIR hyperspectral imaging system. J. Food Eng. 2013, 117, 42–51. [Google Scholar] [CrossRef]
- Xu, S.; Zhao, Y.; Wang, M.; Shi, X. Comparison of multivariate methods for estimating selected soil properties from intact soil cores of paddy fields by Vis–NIR spectroscopy. Geoderma 2018, 310, 29–43. [Google Scholar] [CrossRef]
- Chong, I.-G.; Jun, C.-H. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Afanador, N.L. Important Variable Selection in Partial Least Squares for Industrial Process Understanding and Control. Ph.D. Thesis, Radboud University Nijmegen, Nijmegen, The Netherlands, 2014. [Google Scholar]
- Jung, M.Y.; Lee, M.K.; Park, H.J.; Oh, E.-B.; Shin, J.Y.; Park, J.S.; Jung, S.Y.; Oh, J.-H.; Choi, D.-S. Heat-induced conversion of gingerols to shogaols in ginger as affected by heat type (dry or moist heat), sample type (fresh or dried), temperature and time. Food Sci. Biotechnol. 2017, 27, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Bartley, J.P.; Jacobs, A.L. Effects of drying on flavour compounds in Australian-grown ginger (Zingiber officinale). J. Sci. Food Agric. 2000, 80, 209–215. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samrat, N.H.; Johnson, J.B.; White, S.; Naiker, M.; Brown, P. A Rapid Non-Destructive Hyperspectral Imaging Data Model for the Prediction of Pungent Constituents in Dried Ginger. Foods 2022, 11, 649. https://doi.org/10.3390/foods11050649
Samrat NH, Johnson JB, White S, Naiker M, Brown P. A Rapid Non-Destructive Hyperspectral Imaging Data Model for the Prediction of Pungent Constituents in Dried Ginger. Foods. 2022; 11(5):649. https://doi.org/10.3390/foods11050649
Chicago/Turabian StyleSamrat, Nahidul Hoque, Joel B. Johnson, Simon White, Mani Naiker, and Philip Brown. 2022. "A Rapid Non-Destructive Hyperspectral Imaging Data Model for the Prediction of Pungent Constituents in Dried Ginger" Foods 11, no. 5: 649. https://doi.org/10.3390/foods11050649