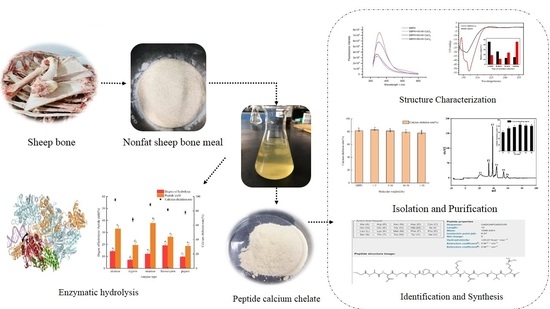
Isolation, Purification and Structure Identification of a Calcium-Binding Peptide from Sheep Bone Protein Hydrolysate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of Sheep Bone Protein Hydrolysate (SBPH)
2.3. Preparation of the Hydrolysate–Calcium Complex
2.4. Amino Acid Analysis
2.5. Characterization of SBPH and SBPH–Ca
2.5.1. Ultraviolet Spectroscopy
2.5.2. Fluorescence Spectroscopy
2.5.3. Zeta Potential
2.5.4. Scanning Electron Microscopy (SEM) and Energy Dispersive Spectroscopy (EDS)
2.5.5. Circular Dichroism (CD) Spectra
2.5.6. Fourier-Transform Infrared Spectroscopy (FTIR)
2.5.7. X-ray Diffraction (XRD)
2.6. Isolation and Purification of Calcium-Binding Peptides
2.6.1. Ultrafiltration
2.6.2. Superdex Peptide Gel Filtration Chromatography
2.6.3. RP-HPLC
2.6.4. Identification of Peptides by Mass Spectrometry
2.6.5. Peptide Synthesis and Validation of Calcium-Chelating Activity
2.7. Statistical Analysis
3. Results and Discussion
3.1. Characteristics of SBPH
3.2. Characterization of SBPH and SBPH–Ca
3.2.1. Ultraviolet Absorption Spectroscopy
3.2.2. Fluorescence Spectroscopy
3.2.3. Zeta Potential
3.2.4. SEM and EDS Analyses
3.2.5. CD Analyses
3.2.6. FTIR Analyses
3.2.7. XRD
3.3. Ultrafiltration
3.4. Separation and Purification of Calcium-Chelating Peptide
3.5. Identification and Synthesis of the Calcium-Binding Peptides
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erfanian, A.; Rasti, B.; Manap, Y. Comparing the calcium bioavailability from two types of nano-sized enriched milk using in-vivo assay. Food Chem. 2017, 214, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Liu, Q.; Bao, X.; Tang, W.; Yang, B.; Guo, S. Identification and characteristics of iron-chelating peptides from soybean protein hydrolysates using IMAC-Fe3+. J. Agric. Food Chem. 2009, 57, 4593–4597. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Silva, M.E.; Netto, F.M.; Bertoldo-Pacheco, M.T.; Alegria, A.; Cilla, A. Peptide-metal complexes: Obtention and role in increasing bioavailability and decreasing the pro-oxidant effect of minerals. Crit. Rev. Food Sci. Nutr. 2021, 61, 1470–1489. [Google Scholar] [CrossRef] [PubMed]
- Cormick, G.; Belizan, J.M. Calcium Intake and Health. Nutrients 2019, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.X.; Wang, X.P.; Guo, X.N. Isolation and characterization of zinc-chelating peptides from wheat germ protein hydrolysates. J. Funct. Foods 2015, 12, 23–32. [Google Scholar] [CrossRef]
- Walters, M.E.; Esfandi, R.; Tsopmo, A. Potential of Food Hydrolyzed Proteins and Peptides to Chelate Iron or Calcium and Enhance their Absorption. Foods 2018, 7, 172. [Google Scholar] [CrossRef]
- Sun, X.; Sarteshnizi, R.A.; Boachie, R.T.; Okagu, O.D.; Abioye, R.O.; Pfeilsticker Neves, R.; Ohanenye, I.C.; Udenigwe, C.C. Peptide-Mineral Complexes: Understanding Their Chemical Interactions, Bioavailability, and Potential Application in Mitigating Micronutrient Deficiency. Foods 2020, 9, 1402. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, Q.; Li, M.; Liu, H.; Wang, Q.; Wu, Y.; Niu, L.; Liu, Z. Isolation of a novel calcium-binding peptide from phosvitin hydrolysates and the study of its calcium chelation mechanism. Food Res. Int. 2021, 141, 110169. [Google Scholar] [CrossRef]
- Liao, W.; Chen, H.; Jin, W.; Yang, Z.; Cao, Y.; Miao, J. Three Newly Isolated Calcium-Chelating Peptides from Tilapia Bone Collagen Hydrolysate Enhance Calcium Absorption Activity in Intestinal Caco-2 Cells. J. Agric. Food Chem. 2020, 68, 2091–2098. [Google Scholar] [CrossRef]
- Malison, A.; Arpanutud, P.; Keeratipibul, S. Chicken foot broth byproduct: A new source for highly effective peptide-calcium chelate. Food Chem. 2021, 345, 128713. [Google Scholar] [CrossRef]
- Wang, L.; Ding, Y.Y.; Zhang, X.X.; Li, Y.F.; Wang, R.; Luo, X.H.; Li, Y.N.; Li, J.; Chen, Z.X. Isolation of a novel calcium-binding peptide from wheat germ protein hydrolysates and the prediction for its mechanism of combination. Food Chem. 2018, 239, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, S.; Zhu, X.; Li, Q.; Fan, Y.; Cheng, D.; Li, B. A novel calcium-binding peptide from Antarctic krill protein hydrolysates and identification of binding sites of calcium-peptide complex. Food Chem. 2018, 243, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Y.-h.; Ma, L.-z.; Kong, B.-h. Hydrolyzing Condition and Immunocompetence of Sheep Bone Protein Enzymatic Lysates. Agric. Sci. China 2009, 8, 1332–1338. [Google Scholar] [CrossRef]
- Cui, P.; Sun, N.; Jiang, P.; Wang, D.; Lin, S. Optimised condition for preparing sea cucumber ovum hydrolysate-calcium complex and its structural analysis. Int. J. Food Sci. Technol. 2017, 52, 1914–1922. [Google Scholar] [CrossRef]
- Cai, X.; Yang, Q.; Lin, J.; Fu, N.; Wang, S. A Specific Peptide with Calcium-Binding Capacity from Defatted Schizochytrium sp. Protein Hydrolysates and the Molecular Properties. Molecules 2017, 22, 544. [Google Scholar] [CrossRef] [PubMed]
- Beyer, R.L.; Hoang, H.N.; Appleton, T.G.; Fairlie, D.P. Metal Clips Induce Folding of a Short Unstructured Peptide into an α-Helix via Turn Conformations in Water. Kinetic versus Thermodynamic Products. J. Am. Chem. Soc. 2004, 126, 15096–15105. [Google Scholar] [CrossRef]
- Sun, N.; Wang, Y.; Bao, Z.; Cui, P.; Wang, S.; Lin, S. Calcium binding to herring egg phosphopeptides: Binding characteristics, conformational structure and intermolecular forces. Food Chem. 2020, 310, 125867. [Google Scholar] [CrossRef]
- Xue, P.; Sun, N.; Li, Y.; Cheng, S.; Lin, S. Targeted regulation of hygroscopicity of soybean antioxidant pentapeptide powder by zinc ions binding to the moisture absorption sites. Food Chem. 2018, 242, 83–90. [Google Scholar] [CrossRef]
- Chang, G.R.; Tu, M.Y.; Chen, Y.H.; Chang, K.Y.; Chen, C.F.; Lai, J.C.; Tung, Y.T.; Chen, H.L.; Fan, H.C.; Chen, C.M. KFP-1, a Novel Calcium-Binding Peptide Isolated from Kefir, Promotes Calcium Influx Through TRPV6 Channels. Mol. Nutr. Food Res. 2021, 65, e2100182. [Google Scholar] [CrossRef]
- Wang, D.; Sun, Z.; Sun, J.; Liu, F.; Du, L.; Wang, D. Preparation and characterization of polylactic acid nanofiber films loading Perilla essential oil for antibacterial packaging of chilled chicken. Int. J. Biol. Macromol. 2021, 192, 379–388. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, L.; Shen, Q.; Qi, L.; Jiang, S.; Guo, Y.; Zhang, C.; Richel, A. Preparation of cattle bone collagen peptides-calcium chelate and its structural characterization and stability. LWT 2021, 144, 111264. [Google Scholar] [CrossRef]
- Vo, T.D.L.; Pham, K.T.; Le, V.M.V.; Lam, H.H.; Huynh, O.N.; Vo, B.C. Evaluation of iron-binding capacity, amino acid composition, functional properties of Acetes japonicus proteolysate and identification of iron-binding peptides. Process Biochem. 2020, 91, 374–386. [Google Scholar] [CrossRef]
- Wu, W.; He, L.; Liang, Y.; Yue, L.; Peng, W.; Jin, G.; Ma, M. Preparation process optimization of pig bone collagen peptide-calcium chelate using response surface methodology and its structural characterization and stability analysis. Food Chem. 2019, 284, 80–89. [Google Scholar] [CrossRef]
- Lin, J.; Cai, X.; Tang, M.; Wang, S. Preparation and Evaluation of the Chelating Nanocomposite Fabricated with Marine Algae Schizochytrium sp. Protein Hydrolysate and Calcium. J. Agric. Food Chem. 2015, 63, 9704–9714. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lan, Y.; Liao, W.; Lin, L.; Liu, G.; Xu, H.; Xue, J.; Guo, B.; Cao, Y.; Miao, J. Preparation, characterization and biological activities of egg white peptides-calcium chelate. LWT 2021, 149, 112035. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, X.; Zhuang, Y. Purification and characterization of novel antioxidant peptides from enzymatic hydrolysates of tilapia (Oreochromis niloticus) skin gelatin. Peptides 2012, 38, 13–21. [Google Scholar] [CrossRef]
- Batoulis, H.; Schmidt, T.H.; Weber, P.; Schloetel, J.G.; Kandt, C.; Lang, T. Concentration Dependent Ion-Protein Interaction Patterns Underlying Protein Oligomerization Behaviours. Sci. Rep. 2016, 6, 24131. [Google Scholar] [CrossRef]
- Cai, X.; Zhao, L.; Wang, S.; Rao, P.J.F. Fabrication and characterization of the nano-composite of whey protein hydrolysate chelated with calcium. Food Funct. 2015, 6, 816–823. [Google Scholar]
- Burstein, E.A.; Vedenkina, N.S.; Ivkova, M.N.J.P. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 2010, 18, 263–279. [Google Scholar] [CrossRef]
- Yang, X.; Yu, X.; Yagoub, A.-G.; Chen, L.; Wahia, H.; Osae, R.; Zhou, C. Structure and stability of low molecular weight collagen peptide (prepared from white carp skin) -calcium complex. LWT 2021, 136, 110335. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Z.; Zhao, Y.; Zeng, M. Enzymatic preparation and characterization of iron-chelating peptides from anchovy (Engraulis japonicus) muscle protein. Food Res. Int. 2012, 48, 435–441. [Google Scholar] [CrossRef]
- Lin, Y.; Cai, X.; Wu, X.; Lin, S.; Wang, S. Fabrication of snapper fish scales protein hydrolysate-calcium complex and the promotion in calcium cellular uptake. J. Funct. Foods 2020, 65, 103717. [Google Scholar] [CrossRef]
- Wang, X.; Gao, A.; Chen, Y.; Zhang, X.; Li, S.; Chen, Y. Preparation of cucumber seed peptide-calcium chelate by liquid state fermentation and its characterization. Food Chem. 2017, 229, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Uppal, R.; Lakshmi, K.V.; Valentine, A.M. Isolation and characterization of the iron-binding properties of a primitive monolobal transferrin from Ciona intestinalis. J. Biol. Inorg. Chem. 2008, 13, 873–885. [Google Scholar] [CrossRef]
- Sun, N.; Jin, Z.; Li, D.; Yin, H.; Lin, S. An Exploration of the Calcium-Binding Mode of Egg White Peptide, Asp-His-Thr-Lys-Glu, and In Vitro Calcium Absorption Studies of Peptide-Calcium Complex. J. Agric. Food Chem. 2017, 65, 9782–9789. [Google Scholar] [CrossRef]
- Guerin, J.; Kriznik, A.; Ramalanjaona, N.; Roux, Y.L.; Girardet, J.M.J. Interaction between dietary bioactive peptides of short length and bile salts in submicellar or micellar state. Food Chem. 2016, 209, 114–122. [Google Scholar] [CrossRef]
- Bao, Z.; Zhang, P.; Sun, N.; Lin, S. Elucidating the Calcium-Binding Site, Absorption Activities, and Thermal Stability of Egg White Peptide-Calcium Chelate. Foods 2021, 10, 2565. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Elavarasan, K.; Shamasundar, B.A.; Badii, F.; Howell, N. Angiotensin I-converting enzyme (ACE) inhibitory activity and structural properties of oven- and freeze-dried protein hydrolysate from fresh water fish (Cirrhinus mrigala). Food Chem. 2016, 206, 210–216. [Google Scholar] [CrossRef]
- Huang, S.L.; Zhao, L.N.; Cai, X.; Wang, S.Y.; Huang, Y.F.; Hong, J.; Rao, P.F. Purification and characterisation of a glutamic acid-containing peptide with calcium-binding capacity from whey protein hydrolysate. J. Dairy Res. 2015, 82, 29–35. [Google Scholar] [CrossRef]
- Barth, A.J.P.i.B.; Biology, M. The infrared absorption of amino acid side chains-ScienceDirect. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef]
- Ye, Q.; Wu, X.; Zhang, X.; Wang, S.J.F. Organic selenium derived from chelation of soybean peptide-selenium and its functional properties in vitro and in vivo. Function 2019, 10, 4761–4770. [Google Scholar] [CrossRef] [PubMed]
- Da, C.; Mu, X.; Hai, H.; Nie, R.; Liu, Z.; Zeng, M. Isolation of a calcium-binding peptide from tilapia scale protein hydrolysate and its calcium bioavailability in rats. J. Funct. Foods 2014, 6, 575–584. [Google Scholar]
- Zhang, K.; Li, B.; Chen, Q.; Zhang, Z.; Zhao, X.; Hou, H. Functional Calcium Binding Peptides from Pacific Cod (Gadus macrocephalus) Bone: Calcium Bioavailability Enhancing Activity and Anti-Osteoporosis Effects in the Ovariectomy-Induced Osteoporosis Rat Model. Nutrients 2018, 10, 091325. [Google Scholar]
- Budseekoad, S.; Yupanqui, C.T.; Sirinupong, N.; Alashi, A.M.; Aluko, R.E.; Youravong, W. Structural and functional characterization of calcium and iron-binding peptides from mung bean protein hydrolysate. J. Funct. Foods 2018, 49, 333–341. [Google Scholar] [CrossRef]
- Caetano-Silva, M.E.; Cilla, A.; Bertoldo-Pacheco, M.T.; Netto, F.M.; Alegría, A. Evaluation of in vitro iron bioavailability in free form and as whey peptide-iron complexes. J. Food Compos. Anal. 2018, 68, 95–100. [Google Scholar] [CrossRef]
- Wang, B.; Xie, N.; Li, B. Influence of peptide characteristics on their stability, intestinal transport, and in vitro bioavailability: A review. J. Food Biochem. 2019, 43, e12571. [Google Scholar] [CrossRef]
- Hou, T.; Wang, C.; Ma, Z.; Shi, W.; Weiwei, L.; He, H. Desalted Duck Egg White Peptides: Promotion of Calcium Uptake and Structure Characterization. J. Agric. Food Chem. 2015, 63, 8170–8176. [Google Scholar] [CrossRef]
- He, J.; Guo, H.; Zhang, M.; Wang, M.; Sun, L.; Zhuang, Y. Purification and Characterization of a Novel Calcium-Binding Heptapeptide from the Hydrolysate of Tilapia Bone with Its Osteogenic Activity. Foods 2022, 11, 468. [Google Scholar] [CrossRef]
- Wu, W.; Li, B.; Hou, H.; Zhang, H.; Zhao, X. Isolation and identification of calcium-chelating peptides from Pacific cod skin gelatin and their binding properties with calcium. Food Funct. 2017, 8, 4441–4448. [Google Scholar] [CrossRef]
- Guo, L.; Harnedy, P.A.; O’Keeffe, M.B.; Zhang, L.; Li, B.; Hou, H.; FitzGerald, R.J. Fractionation and identification of Alaska pollock skin collagen-derived mineral chelating peptides. Food Chem. 2015, 173, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Lin, S.; Han, W.; Jiang, P.; Zhu, B.; Sun, N. The formation mechanism of a sea cucumber ovum derived heptapeptide-calcium nanocomposite and its digestion/absorption behavior. Food Funct. 2019, 10, 8240–8249. [Google Scholar] [CrossRef]
- Lee, S.H. Isolation of a Calcium-binding Peptide from Enzymatic Hydrolysates of Porcine Blood Plasma Protein. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 290–294. [Google Scholar] [CrossRef]
- Liu, F.R.; Wang, L.; Wang, R.; Chen, Z.X. Calcium-binding capacity of wheat germ protein hydrolysate and characterization of Peptide-calcium complex. J. Agric. Food Chem. 2013, 61, 7537–7544. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Liu, S.; Liu, X.; Duan, S.; Xiao, S.; Yang, Z.; Cao, Y.; Miao, J. The purification, identification and bioactivity study of a novel calcium-binding peptide from casein hydrolysate. Food Funct. 2019, 10, 7724–7732. [Google Scholar] [CrossRef] [PubMed]
- Berrocal, R.; Chanton, S.; Juillerat, M.A.; Favillare, B.; Scherz, J.C.; Jost, R. Tryptic phosphopeptides from whole casein. II. Physicochemical properties related to the solubilization of calcium. J. Dairy Res. 1989, 56, 335–341. [Google Scholar] [CrossRef]
- Gravaghi, C.; Favero, E.D.; Cantu’, L.; Donetti, E.; Bedoni, M.; Fiorilli, A.; Tettamanti, G.; Ferraretto, A.J. Casein phosphopeptide promotion of calcium uptake in HT-29 cellsrelationship between biological activity and supramolecular structure. FEBS J. 2007, 19, 274. [Google Scholar]
- Hou, T.; Lu, Y.; Guo, D.; Liu, W.; Shi, W.; He, H. A pivotal peptide (Val-Ser-Glu-Glu) from duck egg white promotes calcium uptake and structure-activity relationship study. J. Funct. Foods 2018, 48, 448–456. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, Y.; Wang, S. Purification of Algal Calcium-Chelating Peptide and Its Physical Chemical Properties. J. Aquat. Food Prod. Technol. 2018, 27, 518–530. [Google Scholar] [CrossRef]
- Gaggelli, E.; D’Amelio, N.; Maccotta, A.; Valensin, G. Calcium-binding properties and molecular organization of bradykinin. Eur. J. Biochem. 1999, 262, 268–276. [Google Scholar] [CrossRef]
- Sun, X.; Ruan, S.; Zhuang, Y.; Sun, L. Anti-osteoporosis effect and purification of peptides with high calcium-binding capacity from walnut protein hydrolysates. Food Funct. 2021, 12, 8454–8466. [Google Scholar] [CrossRef] [PubMed]











| Groups | Time (h) | Temperature (°C) | pH | Protease (U/g) | Calcium Binding Capcity (%) |
|---|---|---|---|---|---|
| A→B | 2→2 | 50 | 9→7.5 | 1:01 | 78.37 ± 0.22 b |
| B→A | 2→2 | 50 | 7.5→9 | 1:01 | 71.54 ± 0.09 d |
| A→C | 2→2 | 50 | 9→7.5 | 1:01 | 81.62 ± 0.32 a |
| C→A | 2→2 | 50 | 7.5→9 | 1:01 | 68.26 ± 0.18 e |
| B→C | 2→2 | 50 | 7.5→7.5 | 1:01 | 76.35 ± 0.25 c |
| C→B | 2→2 | 50 | 7.5→7.5 | 1:01 | 79.18 ± 0.12 b |
| Amino Acid | Content | Amino Acid | Content |
|---|---|---|---|
| Asp | 2.89 ± 0.05 | Lys | 5.04 ± 0.1 |
| Thr | 2.15 ± 0.02 | NH3 | 2.27 ± 0.04 |
| Ser | 2.71 ± 0.06 | His | 2.34 ± 0.04 |
| Glu | 4.76 ± 0.09 | Arg | 8.77 ± 0.23 |
| Gly | 15.68 ± 0.16 | Pro | 6.49 ± 0.15 |
| Ala | 12.31 ± 0.52 | a | 85.21 ± 1.59 |
| Val | 4.11 ± 0.1 | b | 24.66 ± 0.38 |
| Met | 1.47 ± 0.08 | c | 36.26 ± 0.92 |
| Ile | 2.61 ± 0.05 | d | 16.14 ± 0.37 |
| Leu | 5.57 ± 0.13 | e | 7.65 ± 0.14 |
| Tyr | 2.33 ± 0.01 | f | 45.42 ± 0.79 |
| Phe | 3.71 ± 0.06 |
| No. | Sequence | Length | Mass | Leading Razor Protein |
|---|---|---|---|---|
| 1 | HLDDLK | 6 | 739.39 | Q1A2D1 (76–81) |
| 2 | SDLSDL | 6 | 648.30 | Q28745 (82–87) |
| 3 | DLSDLH | 6 | 698.32 | Q28745 (83–88) |
| 4 | HLDDLP | 6 | 708.34 | Q28745 (73–78) |
| 5 | SDLSDLH | 7 | 785.36 | Q28745 (82–88) |
| 6 | HLDDLKG | 7 | 796.41 | Q1A2D1 (76–82] |
| 7 | SLVTGQT | 7 | 704.37 | W5Q638 (145–151) |
| 8 | DFGFDGD | 7 | 771.27 | W5NTT7 (1107–1113) |
| 9 | STGEIGPA | 8 | 730.35 | W5NTT7 (381–388) |
| 10 | FLPQPPQE | 8 | 954.48 | A0A6P7EK74 (1199–1206) |
| 11 | GEAGPQGPR | 9 | 867.42 | A0A6P7EK74 (352–360) |
| 12 | GAPGKDGVRG | 10 | 912.48 | A0A6P7EK74 (754–763) |
| 13 | VEGPPGPEGP | 10 | 934.44 | A0A6P7D8S3 (290–299) |
| 14 | IDGRPGPIGPA | 11 | 1048.57 | W5NTT7 (471–481) |
| 15 | GPAGPPGPIGN | 11 | 932.47 | A0A6P7EK74 (844–854) |
| 16 | SDGSVGPVGPA | 11 | 941.45 | W5NTT7 (234–244) |
| 17 | GADGAPGKDGV | 11 | 942.44 | A0A6P7EK74 (751–761) |
| 18 | DGAPGKDGVRG | 11 | 1027.50 | A0A6P7EK74 (753–763) |
| 19 | GIDGRPGPIGPA | 12 | 1105.59 | W5NTT7 (470–481) |
| 20 | GADGAPGKDGVRG | 13 | 1155.56 | A0A6P7EK74 (751–763) |
| 21 | DFLDEYIFLAVGR | 13 | 1556.79 | A0A6P3TAT2 (393–405) |
| 22 | GPEGPPGEPGPPGPP | 15 | 1337.63 | W5Q4M3 (1209–1223) |
| 23 | STGEAFVQFASQEIAEK | 17 | 1840.88 | A0A6P3YL92 (151–167) |
| No. | Sequence | Mass | Organism | Protein Names | Description | Toxicity | Solubility | Allergenicity | Calcium-Binding Capacity (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GADGAPGKDGVR | 1098.5418 | Bos taurus (Bovine) | Collagen alpha-1(I) chain | Calcium, Metal-binding | NO | Good | NO | 82.25 ± 0.11% |
| 2 | GPSGLPGERG | 925.46174 | Bos taurus (Bovine) | Collagen alpha-2(I) chain | Calcium, Metal-binding | NO | Good | NO | 89.76 ± 0.19% |
| 3 | GAPGKDGVRG | 912.47773 | Bos taurus (Bovine) | Collagen alpha-1(I) chain | Calcium, Metal-binding | NO | Good | NO | 88.26 ± 0.25% |
| 4 | AGPSGPSGLPGERG | 1237.6051 | Bos taurus (Bovine) | Collagen alpha-2(I) chain | Calcium, Metal-binding | NO | Good | NO | 72.97 ± 0.44% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.; Wang, D.; Sun, L.; Su, R.; Corazzin, M.; Sun, X.; Dou, L.; Zhang, M.; Zhao, L.; Su, L.; et al. Isolation, Purification and Structure Identification of a Calcium-Binding Peptide from Sheep Bone Protein Hydrolysate. Foods 2022, 11, 2655. https://doi.org/10.3390/foods11172655
Hu G, Wang D, Sun L, Su R, Corazzin M, Sun X, Dou L, Zhang M, Zhao L, Su L, et al. Isolation, Purification and Structure Identification of a Calcium-Binding Peptide from Sheep Bone Protein Hydrolysate. Foods. 2022; 11(17):2655. https://doi.org/10.3390/foods11172655
Chicago/Turabian StyleHu, Guanhua, Debao Wang, Lina Sun, Rina Su, Mirco Corazzin, Xueying Sun, Lu Dou, Min Zhang, Lihua Zhao, Lin Su, and et al. 2022. "Isolation, Purification and Structure Identification of a Calcium-Binding Peptide from Sheep Bone Protein Hydrolysate" Foods 11, no. 17: 2655. https://doi.org/10.3390/foods11172655