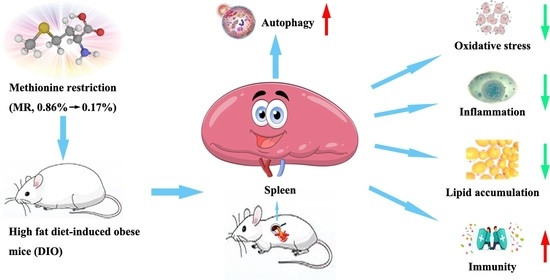
Metabolomics Based on 1H-NMR Reveal the Regulatory Mechanisms of Dietary Methionine Restriction on Splenic Metabolic Dysfunction in Obese Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiment
2.2. Sample Collection
2.3. Analysis of Blood Glucose, Plasma and Spleen Lipid Parameters, and Spleen Crude Protein
2.4. NMR Spectroscopy and Data Processing
2.5. Determination of Oxidative Stress Parameters
2.6. Measurement of Inflammatory Cytokines and Immune Parameters
2.7. Assays for Spleen Glycogen, ATP/ADP/AMP, Mitochondrial DNA Copy Number, and the Respiratory Chain Enzyme Activities
2.8. Gene Expression Assays
2.9. Western Blot Analysis
2.10. Statistical Analysis
3. Results
3.1. MR Normalized Obesity-Induced Changes of Body Weight and Body Composition
3.2. MR Normalized Obesity-Induced Increases in Spleen Weight, Spleen and Plasma Lipid Levels
3.3. MR Normalized Obesity-Induced Changes in the Levels of Spleen Metabolites
3.4. MR Counteracted Obesity-Induced Inhibition in Spleen Energy Production
3.5. MR Reversed Obesity-Induced Enhancement in Spleen Oxidative Stress
3.6. MR Abolished Obesity-Induced Elevation in Spleen Inflammation
3.7. MR Normalized Obesity-Induced Decline in Immune Function
3.8. MR Abolished Obesity-Induced Inhibition in Spleen Autophagy Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bluher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef]
- Akinyemiju, T.; Moore, J.X.; Pisu, M.; Judd, S.E.; Goodman, M.; Shikany, J.M.; Howard, V.J.; Safford, M.; Gilchrist, S.C. A Prospective Study of Obesity, Metabolic Health, and Cancer Mortality. Obesity 2018, 26, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Rius-Perez, S.; Torres-Cuevas, I.; Millan, I.; Ortega, A.L.; Perez, S. PGC-1 alpha, Inflammation, and Oxidative Stress: An Integrative View in Metabolism. Oxid. Med. Cell Longev. 2020, 2020, 1452696. [Google Scholar] [CrossRef] [Green Version]
- Colitti, M.; Stefanon, B.; Gabai, G.; Gelain, M.E.; Bonsembiante, F. Oxidative Stress and Nutraceuticals in the Modulation of the Immune Function: Current Knowledge in Animals of Veterinary Interest. Antioxidants 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lori, A.; Perrotta, M.; Lembo, G.; Carnevale, D. The Spleen: A Hub Connecting Nervous and Immune Systems in Cardiovascular and Metabolic Diseases. Int. J. Mol. Sci. 2017, 18, 1216. [Google Scholar] [CrossRef]
- Wu, L.; Parekh, V.V.; Hsiao, J.; Kitamura, D.; Van Kaer, L. Spleen supports a pool of innate-like B cells in white adipose tissue that protects against obesity-associated insulin resistance. Proc. Natl. Acad. Sci. USA 2014, 111, E4638–E4647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steen, E.H.; Wang, X.Y.; Balaji, S.; Butte, M.J.; Bollyky, P.L.; Keswani, S.G. The Role of the Anti-Inflammatory Cytokine Interleukin-10 in Tissue Fibrosis. Adv. Wound Care 2020, 9, 184–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotoh, K.; Fujiwara, K.; Anai, M.; Okamoto, M.; Masaki, T.; Kakuma, T.; Shibata, H. Role of spleen-derived IL-10 in prevention of systemic low-grade inflammation by obesity. Endocr. J. 2017, 64, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, K.; Inoue, M.; Masaki, T.; Chiba, S.; Shimasaki, T.; Ando, H.; Fujiwara, K.; Katsuragi, I.; Kakuma, T.; Seike, M.; et al. A Novel Anti-inflammatory Role for Spleen-Derived Interleukin-10 in Obesity-Induced Inflammation in White Adipose Tissue and Liver. Diabetes 2012, 61, 1994–2003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wysocki, A.; Drożdż, W.; Dolecki, M. Spleen and lipids metabolism-is there any correlation? Med. Sci. Monit. 1999, 5, CR524–CR527. [Google Scholar]
- Alberti, L.R.; Veloso, D.F.M.; Vasconcellos, L.D.; Petroianu, A. Is there a relationship between lipids metabolism and splenic surgeries? Acta Cir. Bras. 2012, 27, 751–756. [Google Scholar] [CrossRef] [Green Version]
- Ai, X.M.; Ho, L.C.; Han, L.L.; Lu, J.J.; Yue, X.; Yang, N.Y. The role of splenectomy in lipid metabolism and atherosclerosis (AS). Lipids Health Dis. 2018, 17, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Barcena, C.; Quiros, P.M.; Durand, S.; Mayoral, P.; Rodriguez, F.; Caravia, X.M.; Marino, G.; Garabaya, C.; Fernandez-Garcia, M.T.; Kroemer, G.; et al. Methionine Restriction Extends Lifespan in Progeroid Mice and Alters Lipid and Bile Acid Metabolism. Cell Rep. 2018, 24, 2392–2403. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.C.; Kaya, A.; Ma, S.; Kim, G.; Gerashchenko, M.V.; Yim, S.H.; Hu, Z.; Harshman, L.G.; Gladyshev, V.N. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 2014, 5, 3592. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.E.; Johnson, F.B. Methionine Restriction Activates the Retrograde Response and Confers Both Stress Tolerance and Lifespan Extension to Yeast, Mouse and Human Cells. PLoS ONE 2014, 9, e97729. [Google Scholar] [CrossRef] [Green Version]
- Plaisance, E.P.; Greenway, F.L.; Boudreau, A.; Hill, K.L.; Johnson, W.D.; Krajcik, R.A.; Perrone, C.E.; Orentreich, N.; Cefalu, W.T.; Gettys, T.W. Dietary methionine restriction increases fat oxidation in obese adults with metabolic syndrome. J. Clin. Endocrinol. Metab. 2011, 96, E836–E840. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Yang, Y.; Xu, Y.; Gao, Q.; Wu, G.; Jiang, Y.; Sun, J.; Shi, Y.; Le, G. Dietary methionine restriction improves glucose metabolism in the skeletal muscle of obese mice. Food Funct. 2019, 10, 2676–2690. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P., Jr.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Ables, G.P.; Johnson, J.E. Pleiotropic responses to methionine restriction. Exp. Gerontol. 2017, 94, 83–88. [Google Scholar] [CrossRef]
- Tashiro, Y.; Han, Q.; Tan, Y.; Sugisawa, N.; Yamamoto, J.; Nishino, H.; Inubushi, S.; Higuchi, T.; Aoki, T.; Murakami, M.; et al. Oral Recombinant Methioninase Prevents Obesity in Mice on a High-fat Diet. In Vivo 2020, 34, 489–494. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, R.M.; Yang, Z.; Tan, Y.; Han, Q.; Li, S.; Yagi, S. Safety and Toxicity of Recombinant Methioninase and Polyethylene Glycol (PEG) Recombinant Methioninase in Primates. Methods Mol. Biol. 2019, 1866, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Plummer, J.D.; Johnson, J.E. Extension of Cellular Lifespan by Methionine Restriction Involves Alterations in Central Carbon Metabolism and Is Mitophagy-Dependent. Front. Cell Dev. Biol. 2019, 7, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codogno, P.; Meijer, A.J. Autophagy: A Potential Link between Obesity and Insulin Resistance. Cell Metab. 2010, 11, 449–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcena, C.; Lopez-Otin, C.; Kroemer, G. Methionine restriction for improving progeria: Another autophagy-inducing anti-aging strategy? Autophagy 2019, 15, 558–559. [Google Scholar] [CrossRef] [Green Version]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and Metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [Green Version]
- Roca-Agujetas, V.; de Dios, C.; Leston, L.; Mari, M.; Morales, A.; Colell, A. Recent Insights into the Mitochondrial Role in Autophagy and Its Regulation by Oxidative Stress. Oxid. Med. Cell Longev. 2019, 2019, 3809308. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, Y.; Sun, J.; Zhang, J.; Guo, H.; Shi, Y.; Cheng, X.; Tang, X.; Le, G. Dietary methionine restriction reduces hepatic steatosis and oxidative stress in high-fat-fed mice by promoting H2S production. Food Funct. 2019, 10, 61–77. [Google Scholar] [CrossRef]
- Xu, Y.C.; Yang, Y.H.; Sun, J.; Zhang, Y.Y.; Luo, T.Y.; Li, B.W.; Jiang, Y.G.; Shi, Y.H.; Le, G.W. Dietary methionine restriction ameliorates the impairment of learning and memory function induced by obesity in mice. Food Funct. 2019, 10, 1411–1425. [Google Scholar] [CrossRef]
- Yang, Y.H.; Zhang, Y.H.; Xu, Y.C.; Luo, T.Y.; Ge, Y.T.; Jiang, Y.G.; Shi, Y.H.; Sun, J.; Le, G.W. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct. 2019, 10, 5952–5968. [Google Scholar] [CrossRef]
- Ables, G.P.; Perrone, C.E.; Orentreich, D.; Orentreich, N. Methionine-Restricted C57BL/6J Mice Are Resistant to Diet-Induced Obesity and Insulin Resistance but Have Low Bone Density (vol 7, e51357, 2012). PLoS ONE 2014, 9, e104050. [Google Scholar] [CrossRef] [Green Version]
- Orillion, A.R. Dietary Protein (Methionine) Restriction Affects the Tumor Microenvironment by Inducing Macrophage Polarization. State Univ. N. Y. Buffalo 2017, 4, 10273526. [Google Scholar]
- Miller, R.A.; Buehner, G.; Chang, Y.; Harper, J.M.; Sigler, R.; Smith-Wheelock, M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 2005, 4, 119–125. [Google Scholar] [CrossRef]
- Xia, S.F.; Duan, X.M.; Hao, L.Y.; Li, L.T.; Cheng, X.R.; Xie, Z.X.; Qiao, Y.; Li, L.R.; Tang, X.; Shi, Y.H.; et al. Role of thyroid hormone homeostasis in obesity-prone and obesity-resistant mice fed a high-fat diet. Metabolism 2015, 64, 566–579. [Google Scholar] [CrossRef]
- Forney, L.A.; Wanders, D.; Stone, K.P.; Pierse, A.; Gettys, T.W. Concentration-dependent linkage of dietary methionine restriction to the components of its metabolic phenotype. Obesity 2017, 25, 730–738. [Google Scholar] [CrossRef] [Green Version]
- Stone, K.P.; Wanders, D.; Orgeron, M.; Cortez, C.C.; Gettys, T.W. Mechanisms of increased in vivo insulin sensitivity by dietary methionine restriction in mice. Diabetes 2014, 63, 3721–3733. [Google Scholar] [CrossRef] [Green Version]
- Tinsley, F.C.; Taicher, G.Z.; Heiman, M.L. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obes. Res. 2004, 12, 150–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Wang, J.; Li, Y.; Wang, X.; Ren, F.; Wang, X. Structural Studies of Water-Insoluble beta-Glucan from Oat Bran and Its Effect on Improving Lipid Metabolism in Mice Fed High-Fat Diet. Nutrients 2021, 13, 3254. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, P.R.; da Costa, C.A.; de Bem, G.F.; Cordeiro, V.S.; Santos, I.B.; de Carvalho, L.C.; da Conceicao, E.P.; Lisboa, P.C.; Ognibene, D.T.; Sousa, P.J.; et al. Euterpe oleracea Mart.-Derived Polyphenols Protect Mice from Diet-Induced Obesity and Fatty Liver by Regulating Hepatic Lipogenesis and Cholesterol Excretion. PLoS ONE 2015, 10, e0143721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, L.; Dong, F.; Liu, Y.; Li, N.; Li, H.; Lei, H.; Hao, F.; Wang, Y.; Zhu, Y.; et al. Metabonomic Changes Associated with Atherosclerosis Progression for LDLR(-/-) Mice. J. Proteome Res. 2015, 14, 2237–2254. [Google Scholar] [CrossRef] [PubMed]
- Mora-Ortiz, M.; Nunez Ramos, P.; Oregioni, A.; Claus, S.P. NMR metabolomics identifies over 60 biomarkers associated with Type II Diabetes impairment in db/db mice. Metabolomics 2019, 15, 89. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Hu, M.; Liu, M.; Hu, J.Z. (1)H NMR Metabolomics Study of Spleen from C57BL/6 Mice Exposed to Gamma Radiation. Metabolomics 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hu, M.; Feng, J.; Liu, M.; Hu, J.Z. (1)H NMR Metabolomics Study of Metastatic Melanoma in C57BL/6J Mouse Spleen. Metabolomics 2014, 10, 1129–1144. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Tayyari, F.; Khoo, C.; Gu, L.W. A H-1 NMR-based approach to investigate metabolomic differences in the plasma and urine of young women after cranberry juice or apple juice consumption. J. Funct. Foods 2015, 14, 76–86. [Google Scholar] [CrossRef]
- Yang, Y.H.; Zhang, H.; Yan, B.; Zhang, T.Y.; Gao, Y.; Shi, Y.H.; Le, G.W. Health Effects of Dietary Oxidized Tyrosine and Dityrosine Administration in Mice with Nutrimetabolomic Strategies. J. Agric. Food Chem. 2017, 65, 6957–6971. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Liu, L.; Guo, F.; Li, S.; Huang, L.; Sun, C.; Feng, R. Serum metabonomics of NAFLD plus T2DM based on liquid chromatography-mass spectrometry. Clin. Biochem. 2016, 49, 962–966. [Google Scholar] [CrossRef]
- Santacruz, L.; Thomas, O.P.; Duque, C.; Puyana, M.; Tello, E. Comparative Analyses of Metabolomic Fingerprints and Cytotoxic Activities of Soft Corals from the Colombian Caribbean. Mar. Drugs 2019, 17, 37. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Gil-Guzman, E.; Mahran, A.M.; Sharma, R.K.; Nelson, D.R.; Thomas, A.J., Jr.; Agarwa, A. Quality control of reactive oxygen species measurement by luminol-dependent chemiluminescence assay. J. Androl. 2001, 22, 568–574. [Google Scholar] [CrossRef]
- Zhao, F.; Pang, W.T.; Zhang, Z.Y.; Zhao, J.L.; Wang, X.; Liu, Y.; Wang, X.; Feng, Z.H.; Zhang, Y.; Sun, W.Y.; et al. Pomegranate extract and exercise provide additive benefits on improvement of immune function by inhibiting inflammation and oxidative stress in high-fat-diet-induced obesity in rats. J. Nutr. Biochem. 2016, 32, 20–28. [Google Scholar] [CrossRef]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef]
- Pietzke, M.; Arroyo, S.F.; Sumpton, D.; Mackay, G.M.; Martin-Castillo, B.; Camps, J.; Joven, J.; Menendez, J.A.; Vazquez, A. Stratification of cancer and diabetes based on circulating levels of formate and glucose. Cancer Metab. 2019, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.F.; Wang, Z.H.; Zhang, F.F. Association between Related Purine Metabolites and Diabetic Retinopathy in Type 2 Diabetic Patients. Int. J. Endocrinol. 2014, 2014, 651050. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.P.; Wang, C.S.; Liu, F.; Lu, Y.R.; Cheng, J.Q. Metabonomics revealed xanthine oxidase-induced oxidative stress and inflammation in the pathogenesis of diabetic nephropathy. Anal. Bioanal. Chem. 2015, 407, 2569–2579. [Google Scholar] [CrossRef]
- El-Bassossy, H.M.; Watson, M.L. Xanthine oxidase inhibition alleviates the cardiac complications of insulin resistance: Effect on low grade inflammation and the angiotensin system. J. Transl. Med. 2015, 13, 82. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.F.; He, F.; Wu, C.L.; Li, P.; Li, N.Z.; Deng, J.P.; Zhu, G.Q.; Ren, W.K.; Peng, Y.Y. Betaine in inflammation: Mechanistic Aspects and Applications. Front. Immunol. 2018, 9, 1070. [Google Scholar] [CrossRef] [Green Version]
- Marcinkiewicz, J.; Kontny, E. Taurine and inflammatory diseases. Amino Acids 2014, 46, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Park, J. Effects of Black Vinegar and Niacinamide on LPS-Induced Inflammation on Human Keratinocytes. J. Cell. Mol. Physiol. 2020, 3, 193–202. [Google Scholar] [CrossRef]
- Bedhiafi, T.; Charradi, K.; Azaiz, M.B.; Mahmoudi, M.; Msakni, I.; Jebari, K.; Bouziani, A.; Limam, F.; Aouani, E. Supplementation of grape seed and skin extract to orlistat therapy prevents high-fat diet-induced murine spleen lipotoxicity. Appl. Physiol. Nutr. Metab. 2018, 43, 782–794. [Google Scholar] [CrossRef]
- Wang, M.; Xiong, Y.; Zhu, W.; Ruze, R.; Xu, Q.; Yan, Z.; Zhu, J.; Zhong, M.; Cheng, Y.; Hu, S.; et al. Sleeve Gastrectomy Ameliorates Diabetes-Related Spleen Damage by Improving Oxidative Stress Status in Diabetic Obese Rats. Obes. Surg. 2021, 31, 1183–1195. [Google Scholar] [CrossRef]
- Croze, M.L.; Vella, R.E.; Pillon, N.J.; Soula, H.A.; Hadji, L.; Guichardant, M.; Soulage, C.O. Chronic treatment with myo-inositol reduces white adipose tissue accretion and improves insulin sensitivity in female mice. J. Nutr. Biochem. 2013, 24, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, K.; Eaton, S. Mitochondrial beta-oxidation. Eur. J. Biochem. 2004, 271, 462–469. [Google Scholar] [CrossRef]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.H.; Lu, C.Y.; Lee, H.C.; Pang, C.Y.; Ma, Y.S. Oxidative damage and mutation to mitochondrial DNA and age-dependent decline of mitochondrial respiratory function. Ann. N. Y. Acad. Sci. 1998, 854, 155–170. [Google Scholar] [CrossRef]
- de Mello, A.H.; Costa, A.B.; Engel, J.D.G.; Rezin, G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018, 192, 26–32. [Google Scholar] [CrossRef]
- Ikeda, M.; Ide, T.; Fujino, T.; Arai, S.; Saku, K.; Kakino, T.; Tyynismaa, H.; Yamasaki, T.; Yamada, K.; Kang, D.; et al. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS ONE 2015, 10, e0119687. [Google Scholar] [CrossRef]
- Petherick, K.J.; Conway, O.J.L.; Mpamhanga, C.; Osborne, S.A.; Kamal, A.; Saxty, B.; Ganley, I.G. Pharmacological Inhibition of ULK1 Kinase Blocks Mammalian Target of Rapamycin (mTOR)-dependent Autophagy. J. Biol. Chem. 2015, 290, 11376–11383. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xiang, X.H.; Shu, S.S.; Zhang, C.L.; Liang, Y.L.; Jiang, T.T.; Zhang, W.Y.; Guo, T.T.; Liang, X.J.; Tang, X. Advanced oxidation protein products inhibit the autophagy of renal tubular epithelial cells. Exp. Ther. Med. 2018, 15, 3908–3916. [Google Scholar] [CrossRef]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G.; et al. Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification. PLOS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef] [Green Version]
- Kitada, M.; Xu, J.; Ogura, Y.; Monno, I.; Koya, D. Mechanism of Activation of Mechanistic Target of Rapamycin Complex 1 by Methionine. Front. Cell Dev. Biol. 2020, 8, 715. [Google Scholar] [CrossRef]
- Muriach, M.; Flores-Bellver, M.; Romero, F.J.; Barcia, J.M. Diabetes and the Brain: Oxidative Stress, Inflammation, and Autophagy. Oxid. Med. Cell. Longev. 2014, 2014, 102158. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Sowers, J.R.; Ren, J. Targeting autophagy in obesity: From pathophysiology to management. Nat. Rev. Endocrinol. 2018, 14, 356–376. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.J.; Xiang, Y.Q.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010, 24, 3052–3065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papackova, Z.; Dankova, H.; Palenickova, E.; Kazdova, L.; Cahova, M. Effect of Short- and Long-Term High-Fat Feeding on Autophagy Flux and Lysosomal Activity in Rat Liver. Physiol. Res. 2012, 61, S67–S76. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Xu, H.B. Autophagy and Obesity and Diabetes. Adv. Exp. Med. Biol. 2020, 1207, 445–461. [Google Scholar] [CrossRef]
- Wu, M.; Luo, Q.Y.; Nie, R.X.; Yang, X.P.; Tang, Z.Z.; Chen, H. Potential implications of polyphenols on aging considering oxidative stress, inflammation, autophagy, and gut microbiota. Crit. Rev. Food Sci. 2020, 61, 2175–2193. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Bio. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, B.; Korolchuk, V.I.; Sarkar, S. Amino acids and autophagy: Cross-talk and co-operation to control cellular homeostasis. Amino Acids 2015, 47, 2065–2088. [Google Scholar] [CrossRef]
- Yang, Y.H.; Zhang, J.H.; Wu, G.Q.; Sun, J.; Wang, Y.N.; Guo, H.T.; Shi, Y.H.; Cheng, X.R.; Tang, X.; Le, G.W. Dietary methionine restriction regulated energy and protein homeostasis by improving thyroid function in high fat diet mice. Food Funct. 2018, 9, 3718–3731. [Google Scholar] [CrossRef]
- Pasiakos, S.M.; Vislocky, L.M.; Carbone, J.W.; Altieri, N.; Konopelski, K.; Freake, H.C.; Anderson, J.M.; Ferrando, A.A.; Wolfe, R.R.; Rodriguez, N.R. Acute Energy Deprivation Affects Skeletal Muscle Protein Synthesis and Associated Intracellular Signaling Proteins in Physically Active Adults. J. Nutr. 2010, 140, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhou, R.M.; Ma, Z.Y. Autophagy and Energy Metabolism. Autophagy Biol. Dis. Basic Sci. 2019, 1206, 329–357. [Google Scholar] [CrossRef]
- Ueland, P.M.; Midttun, O.; Windelberg, A.; Svardal, A.; Skalevik, R.; Hustad, S. Quantitative profiling of folate and one-carbon metabolism in large-scale epidemiological studies by mass spectrometry. Clin. Chem. Lab. Med. 2007, 45, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.J.; Sanchez-Medina, F.; Torres, M.; Gil, A.; Suarez, A. Dietary nucleotides enhance the liver redox state and protein synthesis in cirrhotic rats. J. Nutr. 2004, 134, 2504–2508. [Google Scholar] [CrossRef] [Green Version]









| Indicators | CON | DIO | DIO + MR |
|---|---|---|---|
| Spleen | |||
| T-AOC (U/mg protein) | 0.86 ± 0.05 | 0.49 ± 0.03 * | 0.83 ± 0.06 # |
| GSH-Px (U/mg protein) | 70.13 ± 2.96 | 51.64 ± 2.33 * | 73.20 ± 3.44 # |
| GSH/GSSG | 1.25 ± 0.06 | 0.89 ± 0.02 * | 1.30 ± 0.05 # |
| ROS (103 cd/mg) | 35.04 ± 1.67 | 57.32 ± 2.45 # | 38.36 ± 2.02 * |
| MDA (nmol/mg protein) | 2.24 ± 0.12 | 3.16 ± 0.10 * | 2.18 ± 0.11 # |
| Plasma | |||
| T-AOC (U/mL) | 3.76 ± 0.17 | 2.57 ± 0.11 * | 3.82 ± 0.20 # |
| GSH-Px (U/mL) | 73.51 ± 3.49 | 51.73 ± 2.85 * | 75.69 ± 4.02 # |
| GSH/GSSG | 0.32 ± 0.02 | 0.25 ± 0.01 * | 0.34 ± 0.02 # |
| ROS (103 cd/μL) | 31.26 ± 2.03 | 46.89 ± 2.69 * | 29.77 ± 2.30 # |
| MDA (nmol/mL) | 13.06 ± 0.81 | 19.22 ± 0.73 * | 14.11 ± 0.71 # |
| Indicators | CON | DIO | DIO + MR |
|---|---|---|---|
| Spleen | |||
| IL-10 (pg/mg protein) | 52.03 ± 2.11 | 34.04 ± 1.55 * | 54.73 ± 2.68 # |
| IL-6 (pg/mg protein) | 32.25 ± 1.30 | 39.12 ± 1.46 * | 33.45 ± 1.19 # |
| IL-1β (pg/mg protein) | 46.81 ± 2.12 | 57.69 ± 1.89 * | 48.33 ± 1.75 # |
| TNF-α (pg/mg protein) | 63.12 ± 2.70 | 84.56 ± 3.23 * | 66.72 ± 2.16 # |
| MCP-1 (pg/mg protein) | 24.66 ± 1.08 | 32.98 ± 1.24 * | 22.21 ± 1.00 # |
| Plasma | |||
| IL-10 (pg/mL) | 145.60 ± 6.52 | 117.14 ± 5.07 * | 138.32 ± 5.86 # |
| IL-6 (pg/mL) | 58.62 ± 1.83 | 73.09 ± 2.35 * | 53.71 ± 1.77 # |
| IL-1β (pg/mL) | 67.53 ± 2.16 | 91.80 ± 2.54 * | 69.64 ± 2.09 # |
| TNF-α (pg/mL) | 116.24 ± 4.05 | 150.67 ± 5.46 * | 121.31 ± 3.13 # |
| MCP-1 (pg/mL) | 36.44 ± 1.20 | 49.79 ± 1.94 * | 37.08 ± 1.69 # |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Qian, J.; Li, B.; Lu, M.; Le, G.; Xie, Y. Metabolomics Based on 1H-NMR Reveal the Regulatory Mechanisms of Dietary Methionine Restriction on Splenic Metabolic Dysfunction in Obese Mice. Foods 2021, 10, 2439. https://doi.org/10.3390/foods10102439
Yang Y, Qian J, Li B, Lu M, Le G, Xie Y. Metabolomics Based on 1H-NMR Reveal the Regulatory Mechanisms of Dietary Methionine Restriction on Splenic Metabolic Dysfunction in Obese Mice. Foods. 2021; 10(10):2439. https://doi.org/10.3390/foods10102439
Chicago/Turabian StyleYang, Yuhui, Jing Qian, Bowen Li, Manman Lu, Guowei Le, and Yanli Xie. 2021. "Metabolomics Based on 1H-NMR Reveal the Regulatory Mechanisms of Dietary Methionine Restriction on Splenic Metabolic Dysfunction in Obese Mice" Foods 10, no. 10: 2439. https://doi.org/10.3390/foods10102439