Machine Learning-Based Automatic Classification of Video Recorded Neonatal Manipulations and Associated Physiological Parameters: A Feasibility Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Setting and Study Sample
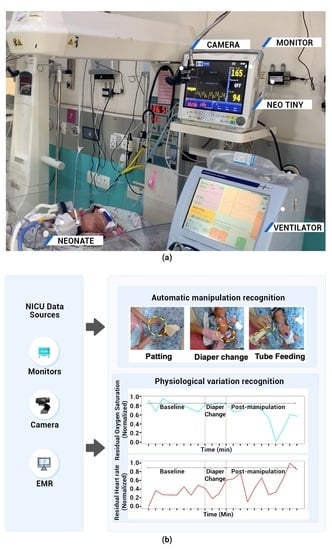
2.2. Data Collection
2.3. Video Acquisition of Manipulation
2.4. Physiological Parameters of Manipulation
2.5. Selection of Manipulations to Be Studied
2.6. Input Data, Training, and Validation Data Set
2.7. Classification of Manipulation Using Convolutional Neural Network (CNN)
2.8. Activity Recognition Combining CNN Output with LSTM
2.9. Variation in Physiological Signals Associated with Manipulation
2.10. Performance Metrics
2.11. Overall Activity Detection Model Evaluation
3. Results
3.1. Baseline Data
3.2. Distribution of Manipulations
3.3. CNN Based Classification of Manipulations
3.4. LSTM Based Classification of Manipulation Videos
3.5. Physiological Signal Variations during Manipulations
- (I)
- For <32 weeks: (a) HR increased during diaper changes and decreased afterward, (b) SpO2 increased during the diaper change.
- (II)
- For ≥32 weeks: (a) HR increased during patting and decreased afterward, (b) the HR decreased after tube feeding.
4. Discussion
5. Limitations
6. Conclusions and Future Directions
7. Code Availability
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Title | Study Done in NICU Population | Video Data | Whether Physiological Data Was Used in the Analysis | Synchronized Video and Physiological Data | Ref |
|---|---|---|---|---|---|
| Monitoring infants by automatic video processing: A unified approach to motion analysis | Yes | Yes | No | No | Cattani et al. [20] |
| Non-contact physiological monitoring of preterm infants in the Neonatal Intensive Care Unit | Yes | Yes | No (vital signs were monitored using video motion analysis of neonates) | No | Villaroel et al. [40] |
| Automatic and continuous discomfort detection for premature infants in a NICU using video-based motion analysis | Yes | Yes | No | No | Sun et al. [41] |
| Multi-Channel Neural Network for Assessing Neonatal Pain from Videos | Yes | Yes | No | No | Salekin et al. [42] |
| Automated pain assessment in neonates | Yes | Yes | Yes (captured from devices using character recognition) | Yes | Zamzmi et al. [43] |
| Intelligent ICU for Autonomous Patient Monitoring Using Pervasive Sensing and Deep Learning | No | Yes | Yes | Yes | Davoudi et al. [44] |
| Machine learning based automatic classification of video recorded neonatal manipulations and associated physiological parameters: A Feasibility Study | Yes | Yes | Yes | Yes | Presented study |
Appendix B




| Characteristics | Details |
|---|---|
| Electrical | |
| Input | 5.0 V, 2 A DC Adaptor (AC 100–240 V, 50/60 Hz) |
| Embedded Battery | LiPo 1 (DC 3.7 V, 1800 mAh) |
| Connectivity | |
| Wired | RS232 × 1 |
| RJ45 × 1 | |
| USB 2 2.0 × 3 | |
| Operating Conditions | |
| Temperature | −20 °C to 70 °C |
| Humidity | 5% to 90% R.H. 3 |
| Memory | 1 GB DDR3 |
| Storage | eMMC: 8 GB |
| CPU | Quad-core 64 bit based on Cortex A53 (4 × 1.5 GHz) |
| Display | 1.8 inch color TFT LCD 4 display (128 × 160 pixel resolution) |
| Dimensions | 77 mm × 58 mm × 41 mm |
| Weight | 150 g |
- [Unit]
- Description = Stream Capturing
- ConditionPathExists = |/usr/bin
- After = network.target
- [Service]
- ExecStart = /usr/local/streampublish/streamPublish.sh
- Restart = always
- RestartSec = 5
- StartLimitInterval = 0
- [Install]
- WantedBy = multi-user.target
- ./capture -F -o -c0 | avconv -re -i - -vcodec libx264 -x264-params keyint = 30:scenecut = 0 -vcodec copy -f mpegts udp://127.0.0.1:1000?pkt_size = 1316
- [Unit]
- Description= Stream publishing to wowza streaming engine
- ConditionPathExists = |/usr/bin
- After = network.target
- [Service]
- ExecStart = /usr/local/srt/srtwrapped.sh
- Restart = always
- RestartSec = 5
- StartLimitInterval = 0
- [Install]
- WantedBy = multi-user.target
- /usr/local/srt/srt-live-transmit udp://127.0.0.1:1000 srt://[wowza server ip]:[port]

| Time Elapsed | Time | Time of Camera | Time of Monitor | Offset (ms) |
|---|---|---|---|---|
| 10 min | Start time | 17:16:58.250 | 17:16:58.205 | 51 |
| End time | 17:26:58.135 | 17:26:58.269 | ||
| 30 min | Start time | 17:39:36.253 | 17:39:36.290 | 27 |
| End time | 18:09:36.211 | 18:09:36.221 | ||
| 60 min | Start time | 18:10:46.308 | 18:10:46.877 | 549 |
| End time | 19:10:46.217 | 19:10:46.237 |
Appendix C



References
- Walani, S.R. Global burden of preterm birth. Int. J. Gynecol. Obstet. 2020, 150, 31–33. [Google Scholar] [CrossRef]
- Kamath, B.D.; Macguire, E.R.; McClure, E.M.; Goldenberg, R.L.; Jobe, A.H. Neonatal Mortality From Respiratory Distress Syndrome: Lessons for Low-Resource Countries. Pediatrics 2011, 127, 1139–1146. [Google Scholar] [CrossRef]
- Koyamaibole, L.; Kado, J.; Qovu, J.D.; Colquhoun, S.; Duke, T. An Evaluation of Bubble-CPAP in a Neonatal Unit in a Developing Country: Effective Respiratory Support That Can Be Applied By Nurses. J. Trop. Pediatr. 2006, 52, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Thukral, A.; Sankar, M.J.; Chandrasekaran, A.; Agarwal, R.; Paul, V.K. Efficacy and safety of CPAP in low- and middle-income countries. J. Perinatol. 2016, 36, S21–S28. [Google Scholar] [CrossRef] [Green Version]
- De Georgia, M.A.; Kaffashi, F.; Jacono, F.J.; Loparo, K.A. Information Technology in Critical Care: Review of Monitoring and Data Acquisition Systems for Patient Care and Research. Sci. World J. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Carayon, P.; Wetterneck, T.B.; Alyousef, B.; Brown, R.L.; Cartmill, R.S.; McGuire, K.; Hoonakker, P.L.T.; Slagle, J.; Van Roy, K.S.; Walker, J.M.; et al. Impact of electronic health record technology on the work and workflow of physicians in the intensive care unit. Int. J. Med. Inform. 2015, 84, 578–594. [Google Scholar] [CrossRef] [Green Version]
- Mark, R. The Story of MIMIC. In Secondary Analysis of Electronic Health Records; Secondary Analysis of Electronic Health Records; Data, M.C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 43–49. [Google Scholar]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harutyunyan, H.; Khachatrian, H.; Kale, D.C.; Ver Steeg, G.; Galstyan, A. Multitask learning and benchmarking with clinical time series data. Sci. Data 2019, 6, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairchild, K.D.; Lake, D.E.; Kattwinkel, J.; Moorman, J.R.; Bateman, D.A.; Grieve, P.G.; Isler, J.R.; Sahni, R. Vital signs and their cross-correlation in sepsis and NEC: A study of 1,065 very-low-birth-weight infants in two NICUs. Pediatr. Res. 2017, 81, 315. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, K.D.; Sinkin, R.A.; Davalian, F.; Blackman, A.E.; Swanson, J.R.; Matsumoto, J.A.; Lake, D.E.; Moorman, J.R.; Blackman, J.A. Abnormal heart rate characteristics are associated with abnormal neuroimaging and outcomes in extremely low birth weight infants. J. Perinatol. 2014, 34, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Saria, S.; Rajani, A.K.; Gould, J.; Koller, D.; Penn, A.A. Integration of Early Physiological Responses Predicts Later Illness Severity in Preterm Infants. Sci. Transl. Med. 2010, 2, 48ra65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairchild, K. Aschner HeRO monitoring to reduce mortality in NICU patients. RRN 2012, 2, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Jeba, A.; Kumar, S. Shivaprakash sosale Effect of positioning on physiological parameters on low birth weight preterm babies in neonatal intensive care unit. Int. J. Res. Pharm. Sci. 2019, 10, 2800–2804. [Google Scholar] [CrossRef]
- Barbosa, A.L.; Cardoso, M.V.L.M.L. Alterations in the physiological parameters of newborns using oxygen therapy in the collection of blood gases. Acta Paul. Enferm. 2014, 4, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Catelin, C.; Tordjman, S.; Morin, V.; Oger, E.; Sizun, J. Clinical, Physiologic, and Biologic Impact of Environmental and Behavioral Interventions in Neonates during a Routine Nursing Procedure. J. Pain 2005, 6, 791–797. [Google Scholar] [CrossRef]
- Pereira, F.L.; Góes, F.; Fonseca, L.M.M.; Scochi, C.G.S.; Castral, T.C.; Leite, A.M. Handling of preterm infants in a neonatal intensive care unit. Rev. Esc. Enferm. USP 2013, 47, 1272–1278. [Google Scholar] [CrossRef] [Green Version]
- Ellsworth, M.A.; Lang, T.R.; Pickering, B.W.; Herasevich, V. Clinical data needs in the neonatal intensive care unit electronic medical record. BMC Med. Inform. Decis. Mak. 2014, 14, 92. [Google Scholar] [CrossRef] [Green Version]
- Moccia, S.; Migliorelli, L.; Carnielli, V.; Frontoni, E. 2019 Preterm infants’ pose estimation with spatio-temporal features. IEEE Trans. Biomed. Eng. 2020, 67, 2370–2380. [Google Scholar] [CrossRef]
- Cattani, L.; Alinovi, D.; Ferrari, G.; Raheli, R.; Pavlidis, E.; Spagnoli, C.; Pisani, F. Monitoring infants by automatic video processing: A unified approach to motion analysis. Comput. Biol. Med. 2017, 80, 158–165. [Google Scholar] [CrossRef]
- Sizun, J.; Ansquer, H.; Browne, J.; Tordjman, S.; Morin, J.-F. Developmental care decreases physiologic and behavioral pain expression in preterm neonates. J. Pain 2002, 3, 446–450. [Google Scholar] [CrossRef]
- Solberg, S.; Morse, J.M. The Comforting Behaviors of Caregivers toward Distressed Postoperative Neonates. Issues Compr. Pediatr. Nurs. 1991, 14, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Chrupcala, K.A.; Edwards, T.M.; Spatz, D.L. A Continuous Quality Improvement Project to Implement Infant-Driven Feeding as a Standard of Practice in the Newborn/Infant Intensive Care Unit. J. Obstet. Gynecol. Neonatal Nurs. 2015, 44, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Kirk, A.T.; Alder, S.C.; King, J.D. Cue-based oral feeding clinical pathway results in earlier attainment of full oral feeding in premature infants. J. Perinatol. 2007, 27, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Yadav, G.; Mallaiah, R.; Joshi, P.; Joshi, V.; Kaur, R.; Bansal, S.; Brahmachari, S.K. iNICU—Integrated Neonatal Care Unit: Capturing Neonatal Journey in an Intelligent Data Way. J. Med. Syst. 2017, 41, 132. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Kaur, R.; Gangadharan, A.; Pandey, A.K.; Manur, A.; Sun, Y.; Saluja, S.; Gupta, S.; Palma, J.P.; Kumar, P. Neo-Bedside Monitoring Device for Integrated Neonatal Intensive Care Unit (iNICU). IEEE Access 2019, 7, 7803–7813. [Google Scholar] [CrossRef]
- Comaru, T.; Miura, E. Postural support improves distress and pain during diaper change in preterm infants. J. Perinatol. 2009, 29, 504–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-W.; Chang, Y.-J. A Preliminary Study of Bottom Care Effects on Premature Infants’ Heart Rate and Oxygen Saturation. J. Nurs. Res. 2004, 12, 161–168. [Google Scholar] [CrossRef]
- Jadcherla, S.R.; Chan, C.Y.; Moore, R.; Malkar, M.; Timan, C.J.; Valentine, C.J. Impact of feeding strategies on the frequency and clearance of acid and nonacid gastroesophageal reflux events in dysphagic neonates. J. Parenter. Enter. Nutr. 2012, 36, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, USA, 27–30 June 2016; 2016; pp. 2818–2826. [Google Scholar]
- Shao, L.; Zhu, F.; Li, X. Transfer Learning for Visual Categorization: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2015, 26, 1019–1034. [Google Scholar] [CrossRef]
- Wharton, Z.; Thomas, E.; Debnath, B.; Behera, A. A vision-based transfer learning approach for recognizing behavioral symptoms in people with dementia. In Proceedings of the 2018 15th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS), Auckland, New Zealand, 27–30 November 2018; pp. 1–6. [Google Scholar]
- Maaten, L.V.D.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Gulli, A.; Pal, S. Deep learning with Keras; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A System for Large-Scale Machine Learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ’16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Prechelt, L. Early Stopping—But When? In Neural Networks: Tricks of the Trade; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1998; Volume 1524, pp. 55–69. [Google Scholar]
- Hall, R.W.; Anand, K.J. Pain management in newborns. Clin. Perinatol. 2014, 41, 895–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Profit, J.; Gould, J.B.; Bennett, M.; Goldstein, B.A.; Draper, D.; Phibbs, C.S.; Lee, H.C. Racial/Ethnic Disparity in NICU Quality of Care Delivery. Pediatrics 2017, 140, e20170918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horbar, J.D.; Edwards, E.M.; Greenberg, L.T.; Profit, J.; Draper, D.; Helkey, D.; Lorch, S.A.; Lee, H.C.; Phibbs, C.S.; Rogowski, J.; et al. Racial Segregation and Inequality in the Neonatal Intensive Care Unit for Very Low-Birth-Weight and Very Preterm Infants. JAMA Pediatr. 2019, 173, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, M.; Chaichulee, S.; Jorge, J.; Davis, S.; Green, G.; Arteta, C.; Zisserman, A.; McCormick, K.; Watkinson, P.; Tarassenko, L. Non-contact physiological monitoring of preterm infants in the Neonatal Intensive Care Unit. NPJ Digit. Med. 2019, 2, 128. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Kommers, D.; Wang, W.; Joshi, R.; Shan, C.; Tan, T.; Aarts, R.M.; van Pul, C.; Andriessen, P.; de With, P.H.N. Automatic and Continuous Discomfort Detection for Premature Infants in a NICU Using Video-Based Motion Analysis. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 5995–5999. [Google Scholar]
- Salekin, M.S.; Zamzmi, G.; Goldgof, D.; Kasturi, R.; Ho, T.; Sun, Y. Multi-Channel Neural Network for Assessing Neonatal Pain from Videos. In Proceedings of the 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC), Bari, Italy, 6–9 October 2019; pp. 1551–1556. [Google Scholar]
- Zamzmi, G.; Pai, C.-Y.; Goldgof, D.; Kasturi, R.; Sun, Y.; Ashmeade, T. Automated pain assessment in neonates. In Scandinavian Conference on Image Analysis; Springer: Cham, Switzerland, 2017; pp. 350–361. [Google Scholar]
- Davoudi, A.; Malhotra, K.R.; Shickel, B.; Siegel, S.; Williams, S.; Ruppert, M.; Bihorac, E.; Ozrazgat-Baslanti, T.; Tighe, P.J.; Bihorac, A.; et al. Intelligent ICU for Autonomous Patient Monitoring Using Pervasive Sensing and Deep Learning. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Linux, T.V. Available online: https://linuxtv.org/downloads/legacy/video4linux/v4l2dwgNew.html (accessed on 21 May 2020).
- Wowza Streaming Engine. Available online: https://www.wowza.com/docs/wowza-streaming-engine-product-articles (accessed on 14 January 2020).
- Zaidi, S.; Bitam, S.; Mellouk, A. Enhanced user datagram protocol for video streaming in VANET. In Proceedings of the 2017 IEEE International Conference on Communications (ICC), Paris, France, 21–25 May 2017; 2017; pp. 1–6. [Google Scholar]
- SRT Alliance. Available online: https://www.srtalliance.org/ (accessed on 14 January 2020).
- Ruether, T. Wowza Product Resources Center. Available online: https://www.wowza.com/blog/streaming-protocols (accessed on 29 January 2020).
- Record Live Video to VOD. Available online: https://www.wowza.com/docs/how-to-record-live-streams-wowza-streaming-engine#record-all-incoming-streams (accessed on 13 April 2020).
- WebRTC. Available online: https://webrtc.org (accessed on 19 February 2020).
- Highcharts. Available online: https://www.highcharts.com (accessed on 10 February 2020).
- Bender, D.; Sartipi, K. HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In Proceedings of the 26th IEEE international symposium on computer-based medical systems, Porto, Portugal, 20–22 June 2013; 2013; pp. 326–331. [Google Scholar]
- ASTM International. Available online: https://www.astm.org/ (accessed on 22 May 2020).








| Manipulation: | Characteristics | Ref |
|---|---|---|
| Patting: | Definition: This is a comforting manipulation where the flat surface of the palmer surface of the caregiver’s hand was brought into contact with a surface of the neonate’s body singly or repetitively. The intensity and rate were variable in different episodes of patting. | [22] |
| Spatial features: Nurse’s hand, neonate’s body boundaries | ||
| Temporal features: Frequency: On-demand Duration: 33 s | ||
| Diaper Change: | Definition: This manipulation involves changing the diaper and cleaning the diaper area for skin hygiene. | [27,28] |
| Spatial features: Two nurse’s hands, diaper, and skin contrast | ||
| Temporal features: Frequency: 4 h Duration: 3 min | ||
| Tube Feeding: | Definition: This manipulation utilizes a soft tube placed through the nose (nasogastric) or mouth (orogastric) placed into the stomach. The feeding is provided through a tube into the stomach until the baby can take food by mouth. | [29] |
| Spatial features: Nurse’s hand, milk, syringe attached to the feeding tube (with or without plunger) | ||
| Temporal features: Frequency: 2 h Duration: 10–30 min |
| Id | Sex | Gestational Age | Birth Weight (g) | Age Interval for Recording (Days) | Clinical Diagnoses |
|---|---|---|---|---|---|
| 1 | Male | 26+0 | 1005 | 24–25 | RDS, Apnea, Prematurity |
| 2 | Male | 27+1 | 800 | 76–90 | Prematurity |
| 3 | Male | 29+4 | 1372 | 37–44 | Prematurity, RDS, Apnea Sepsis |
| 4 | Male | 35+2 | 1400 | 8–10 | NNH |
| 5 | Male | 36+0 | 2400 | 3–5 | RDS, NNH |
| 6 | Male | 36+6 | 1430 | 4–8 | Prematurity, NNH |
| 7 | Male | 36+6 | 3231 | 5–6 | RDS |
| 8 | Male | 39+2 | 2600 | 7–8 | RDS, Seizure |
| 9 | Male | 39+4 | 2000 | 5–6 | Sepsis, RDS, Apnea |
| 10 | Male | 40+0 | 2700 | 3–7 | RDS, NNH |
| Manipulation | # Frequency | * Average Duration (Seconds) | Minimum Duration (Seconds) | Maximum Duration (Seconds) |
|---|---|---|---|---|
| Patting | 167 | 28.9 (12.4) | 12 | 56 |
| Tube Feeding | 108 | 108.9 (55.3) | 25 | 300 |
| Diaper Change | 64 | 45.5 (18.8) | 17 | 92 |
| Patting | Nurse | Not Captured in EMR |
| NTS | The patting was started at 14:05:08 on 17-08-2020 and completed at 14:06:19 (duration: 71 s). This is manipulation number 3, since 8 a.m. | |
| Diaper Change | Nurse | Not captured in EMR |
| NTS | The diaper change was started at 19:35:25 on 17-08-2020 and completed at 19:37:01 (duration: 96 s). This is manipulation number 4 since 8 a.m. | |
| Tube feed Entry | Nurse | Start Time: 17-08-2020 09:30 a.m. Type: Tube Feed Type of Milk: Preterm Formula Quantity: 11 mL |
| NTS | The feeding was started at 09:30:09 on 17-08-2020 and completed at 09:32:57 (duration: 168 s). This is manipulation number 1 since 8 a.m. |
| PPV | Sensitivity | F-Measure | Total Manipulations | |
|---|---|---|---|---|
| Patting | 0.86 | 1.00 | 0.92 | 167 |
| Diaper Change | 0.98 | 0.68 | 0.80 | 64 |
| Tube feeding | 1.00 | 0.87 | 0.93 | 108 |
| <32 Weeks | ≥32 Weeks | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Manipulations | Parameters | Baseline * | During * | Post * | p-Value $ | p-Value # | Baseline * | During * | Post * | p-Value $ | p-Value # |
| Patting | HR (BPM) | 161.9 (10.19) | 164.7 (13.7) | 157.6 (24.9) | 0.168 | 0.069 | 148.7 (13.9) | 165.7 (30.7) | 150.9 (8.2) | 0.019 | 0.00 |
| SpO2 (%) | 92.7 (7.4) | 93.0 (7.9) | 89.7 (12.8) | 0.43 | 0.087 | 94.7 (6.1) | 92.5 (10.9) | 93.5 (11.41) | 0.21 | 0.34 | |
| Diaper Change | HR (BPM) | 152.738 (31.4) | 166.9 (14.4) | 157.4 (23.2) | 0.000 | 0.036 | 147.8 (12.02) | 152.7 (15.8) | 150.7 (9.5) | 0.10 | 0.17 |
| SpO2 (%) | 88.9 (18.2) | 94.02 (5.7) | 89.4 (13.6) | 0.000 | 0.07 | 94.7 (5.8) | 94.9 (5.4) | 93.9 (12.7) | 0.44 | 0.36 | |
| Tube Feeding | HR (BPM) | 163.1 (10.55) | 164.28 (13.29) | 162.2 (20.0) | 0.26 | 0.22 | 150.5 (16.7) | 147.6 (16.6) | 153.3 (11.6) | 0.17 | 0.003 |
| SpO2 (%) | 93.9 (6.4) | 93.9 (4.9) | 91.7 (9.9) | 0.49 | 0.052 | 95.1 (4.9) | 94.0 (8.0) | 93.5 (7.6) | 0.23 | 0.37 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, H.; Kusuda, S.; McAdams, R.M.; Gupta, S.; Kalra, J.; Kaur, R.; Das, R.; Anand, S.; Pandey, A.K.; Cho, S.J.; et al. Machine Learning-Based Automatic Classification of Video Recorded Neonatal Manipulations and Associated Physiological Parameters: A Feasibility Study. Children 2021, 8, 1. https://doi.org/10.3390/children8010001
Singh H, Kusuda S, McAdams RM, Gupta S, Kalra J, Kaur R, Das R, Anand S, Pandey AK, Cho SJ, et al. Machine Learning-Based Automatic Classification of Video Recorded Neonatal Manipulations and Associated Physiological Parameters: A Feasibility Study. Children. 2021; 8(1):1. https://doi.org/10.3390/children8010001
Chicago/Turabian StyleSingh, Harpreet, Satoshi Kusuda, Ryan M. McAdams, Shubham Gupta, Jayant Kalra, Ravneet Kaur, Ritu Das, Saket Anand, Ashish Kumar Pandey, Su Jin Cho, and et al. 2021. "Machine Learning-Based Automatic Classification of Video Recorded Neonatal Manipulations and Associated Physiological Parameters: A Feasibility Study" Children 8, no. 1: 1. https://doi.org/10.3390/children8010001