Microbiome in Nasal Mucosa of Children and Adolescents with Allergic Rhinitis: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Study Selection
2.4. Data Extraction
2.5. Study Quality Assessment
3. Results
3.1. Number of Retrieved Papers
3.2. Microbiome Diversity and Richness
3.3. Taxonomic Composition
3.3.1. Nasal Cavity Microbiota
3.3.2. Nasopharyngeal Microbiota
3.3.3. Hypopharyngeal Microbiota
4. Discussion
5. Conclusions
- (1)
- ethnical records of the individuals;
- (2)
- allergic predisposition records;
- (3)
- environmental factors;
- (4)
- pharmacotherapy used;
- (5)
- anatomical variants (e.g., adenotonsillar hypertrophy or nasal septal deviation).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Losol, P.; Park, H.S.; Song, W.J.; Hwang, Y.K.; Kim, S.H.; Holloway, J.W.; Chang, Y.S. Association of upper airway bacterial microbiota and asthma: Systematic review. Asia Pac. Allergy 2022, 12, e32. [Google Scholar] [CrossRef]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Goulding, D.; Stabler, R.A.; Croucher, N.; Mastroeni, P.; Scott, P.; Raisen, C.; Mottram, L.; et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009, 77, 3661–3669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.K.; Huang, Y.J.; LiPuma, J.J.; Boushey, H.A.; Boucher, R.C.; Cookson, W.O.; Curtis, J.L.; Erb-Downward, J.; Lynch, S.V.; Sethi, S.; et al. Significance of the microbiome in obstructive lung disease. Thorax 2012, 67, 456–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitri-Pinheiro, S.; Soares, R.; Barata, P. The microbiome of the nose—Friend or foe? Allergy Rhinol. 2020, 11, 2152656720911605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, J.; Han, M.S.; Kwak, J.; Kim, T.H. Association Between Microbiota and Nasal Mucosal Diseases in terms of Immunity. Int. J. Mol. Sci. 2021, 22, 4744. [Google Scholar] [CrossRef]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome composition and its impact on the development of allergic diseases. Front. Immun. 2020, 11, 700. [Google Scholar] [CrossRef] [Green Version]
- Gan, W.; Yang, F.; Meng, J.; Liu, F.; Liu, S.; Xian, J. Comparing the nasal bacterial microbiome diversity of allergic rhinitis, chronic rhinosinusitis and control subjects. Eur. Arch. Otorhinolaryngol. 2021, 278, 711–718. [Google Scholar] [CrossRef]
- Brindisi, G.; Marazzato, M.; Brunetti, F.; De Castro, G.; Loffredo, L.; Carnevale, R.; Cinicola, B.; Palamara, A.T.; Conte, M.P.; Zicari, A.M. Allergic rhinitis, microbiota and passive smoke in children: A pilot study. Pediatr. Allergy Immunol. 2022, 33, 22–26. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Lee, T.D.; Andrade, J.; Oguntuyo, K.; Chun, Y.; Grishina, G.; Grishin, A.; Bunyavanich, S. The nasal microbiome in asthma. J. Allergy Clin. Immunol. 2018, 142, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Mahdavinia, M. The nasal microbiome: Opening new clinical research avenues for allergic disease. Expert Rev. Clin. Immunol. 2018, 14, 645–647. [Google Scholar] [CrossRef] [Green Version]
- Chiang, T.Y.; Yang, Y.R.; Zhuo, M.Y.; Yang, F.; Zhang, Y.F.; Fu, C.H.; Lee, T.J.; Chung, W.H.; Chen, L.; Chang, C.J. Microbiome profiling of nasal extracellular vesicles in patients with allergic rhinitis. World Allergy Organ. J. 2022, 15, 100674. [Google Scholar] [CrossRef]
- Marazzato, M.; Zicari, A.M.; Aleandri, M.; Conte, A.L.; Longhi, C.; Vitanza, L.; Bolognino, V.; Zagaglia, C.; De Castro, G.; Brindisi, G.; et al. 16S metagenomics reveals dysbiosis of nasal core microbiota in children with chronic nasal inflammation: Role of adenoid hypertrophy and allergic rhinitis. Front. Cell. Infect. Microbiol. 2020, 10, 458. [Google Scholar] [CrossRef]
- Lee, J.T.; Kim, C.M.; Ramakrishnan, V. Microbiome and disease in the upper airway. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 1–6. [Google Scholar] [CrossRef]
- Yau, J.W.K.; Hou, J.; Tsui, S.K.W.; Leung, T.F.; Cheng, N.S.; Yam, J.C.; Kam, K.W.; Jhanji, V.; Hon, K.L. Characterization of ocular and nasopharyngeal microbiome in allergic rhinoconjunctivitis. Pediatr. Allergy Immunol. 2019, 30, 624–631. [Google Scholar] [CrossRef]
- Liu, T.; Lin, C.H.; Chen, Y.L.; Jeng, S.L.; Tsai, H.J.; Ho, C.L.; Kuo, W.S.; Hsieh, M.H.; Chen, P.C.; Wu, L.S.; et al. Nasal Microbiome Change During and After Exacerbation in Asthmatic Children. Front. Microbiol. 2021, 12, 833726. [Google Scholar] [CrossRef]
- Zhou, Y.; Jackson, D.; Bacharier, L.B.; Mauger, D.; Boushey, H.; Castro, M.; Durack, J.; Huang, Y.; Lemanske, R.F.; Storch, G.A.; et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat. Commun. 2019, 10, 5714. [Google Scholar] [CrossRef] [Green Version]
- Morin, A.; McKennan, C.G.; Pedersen, C.E.T.; Stokholm, J.; Chawes, B.L.; Schoos, A.M.M.; Naughton, K.A.; Thorsen, J.; Mortensen, M.S.; Vercelli, D.; et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J. Allergy Clin. Immunol. 2020, 146, 1358–1366. [Google Scholar] [CrossRef]
- Ta, L.D.H.; Yap, G.C.; Tay, C.J.X.; Lim, A.S.M.; Huang, C.H.; Chu, C.W.; De Sessions, P.F.; Shek, L.P.; Goh, A.; Van Bever, H.P.; et al. Establishment of the nasal microbiota in the first 18 months of life: Correlation with early-onset rhinitis and wheezing. J. Allergy Clin. Immunol. 2018, 142, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.Y.; Chan, Y.L.; Tsai, Y.S.; Chen, S.A.; Wang, C.J.; Chen, K.F.; Chung, I. Airway microbial diversity is inversely associated with mite-sensitized rhinitis and asthma in early childhood. Sci. Rep. 2017, 7, 1820. [Google Scholar] [CrossRef] [Green Version]
- Bisgaard, H.; Hermansen, M.N.; Buchvald, F.; Loland, L.; Halkjaer, L.B.; Bønnelykke, K.; Brasholt, M.; Heltberg, A.; Vissing, N.H.; Thorsen, S.V.; et al. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007, 357, 1487–1495. [Google Scholar] [CrossRef]
- Chen, M.; He, S.; Miles, P.; Li, C.; Ge, Y.; Yu, X.; Wang, L.; Huang, W.; Kong, X.; Ma, S.; et al. Nasal Bacterial Microbiome Differs between Healthy Controls and Those with Asthma and Allergic Rhinitis. Front. Cell. Infect. Microbiol. 2022, 12, 841995. [Google Scholar] [CrossRef] [PubMed]
- Scadding, G.K.; Smith, P.K.; Blaiss, M.; Roberts, G.; Hellings, P.W.; Gevaert, P.; Mc Donald, M.; Sih, T.; Halken, S.; Zieglmayer, P.U.; et al. Allergic Rhinitis in Childhood and the New EUFOREA Algorithm. Front. Allergy 2021, 2, 706589. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Heart Lung and Blood Institute. Study Quality Assessment Tools; NHLBI: Bethesda, MD, USA, 2019.
- Sterne, J.A.C.; Savovic, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. Br. Med. J. 2019, 366, l4898. [Google Scholar] [CrossRef] [Green Version]
- Amos, G.C.; Logan, A.; Anwar, S.; Fritzsche, M.; Mate, R.; Bleazard, T.; Rijpkema, S. Developing standards for the microbiome field. Microbiome 2020, 8, 98. [Google Scholar] [CrossRef]
- Augustine, T.; Kumar, M.; Al Khodor, S.; van Panhuys, N. Microbial Dysbiosis Tunes the Immune Response towards Allergic Disease Outcomes. Clin. Rev. Allergy Immunol. 2022, 1–29. [Google Scholar] [CrossRef]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87. [Google Scholar] [CrossRef] [Green Version]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Lappan, R.; Jamieson, S.E.; Peacock, C.S. Reviewing the pathogenic potential of the otitis-associated Bacteria Alloiococcus otitidis and Turicella otitidis. Front. Cell. Infect. Microbiol. 2020, 10, 51. [Google Scholar] [CrossRef]
- Kezele, G.T.; Abram, M.; Bubonja-Šonje, M. Alloiococcus otitidis—Cause of nonspecific acute sinusitis: First case report and review of Literature. Microorganisms 2022, 10, 1182. [Google Scholar] [CrossRef]
- Folino, F.; Fattizzo, M.; Ruggiero, L.; Oriano, M.; Aliberti, S.; Blasi, F.; Gaffuri, M.; Marchisio, P. Nasopharyngeal microbiota analysis in healthy and otitis-prone children: Focus on history of spontaneous tympanic membrane perforation. J. Pediatr. Infect. Dis. 2020, 40, 16–21. [Google Scholar] [CrossRef]
- Montaño-Velázquez, B.B.; Navarrete, R.C.; Mogica Martínez, M.D.; Becerril-Ángeles, M.; Jáuregui-Renaud, K. Rhinomanometry in young patients with perennial allergic rhinitis with/without recent exposure to tobacco smoke. Clin. Otolaryngol. 2011, 36, 320–324. [Google Scholar] [CrossRef]
- Santee, C.A.; Nagalingam, N.A.; Faruqi, A.A.; DeMuri, G.P.; Gern, J.E.; Wald, E.R.; Lynch, S.V. Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome 2016, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Vissing, N.H.; Chawes, B.L.; Bisgaard, H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am. J. Respir. Crit. Care Med. 2013, 188, 1246–1252. [Google Scholar] [CrossRef]
- Shargorodsky, J.; Garcia-Esquinas, E.; Navas-Acien, A.; Lin, S.Y. Allergic sensitization, rhinitis, and tobacco smoke exposure in US children and adolescents. Int. Forum Allergy Rhinol. 2015, 5, 471–476. [Google Scholar] [CrossRef] [Green Version]
- Yao, T.C.; Chang, S.W.; Chang, W.C.; Tsai, M.H.; Liao, S.L.; Hua, M.C.; Lai, S.H.; Yeh, K.W.; Tseng, Y.L.; Lin, W.C.; et al. Exposure to tobacco smoke and childhood rhinitis: A population-based study. Sci. Rep. 2017, 7, 42836. [Google Scholar] [CrossRef] [Green Version]
- Saulyte, J.; Regueira, C.; Montes-Martínez, A.; Khudyakov, P.; Takkouche, B. Active or passive exposure to tobacco smoking and allergic rhinitis, allergic dermatitis, and food allergy in adults and children: A systematic review and meta-analysis. PLoS Med. 2014, 11, 1001611. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Ma, Y.; Xu, T.; Zhang, Q.Z.; Bai, J.; Wang, J.; Zhu, T.; Lou, Q.; Götz, F.; Qu, D.; et al. Nicotine enhances Staphylococcus epidermidis biofilm formation by altering the bacterial autolysis, extracellular DNA releasing, and polysaccharide intercellular adhesin production. Front. Microbiol. 2018, 9, 2575. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Han, S.A.; Kim, W. Compositional alterations of the nasal microbiome and Staphylococcus aureus—Characterized dysbiosis in the nasal mucosa of patients with allergic rhinitis. Clin. Exp. Otorhinolaryngol. 2022, 15, 335–345. [Google Scholar] [CrossRef]
- Agarwal, A.; Kaushik, A.; Kumar, S.; Mishra, R.K. Comparative study on air quality status in Indian and Chinese cities before and during the COVID-19 lockdown period. Air Qual. Atmos. Health 2020, 13, 1167–1178. [Google Scholar] [CrossRef]
- Baptista, E.A.; Dey, S.; Pal, S. Chronic respiratory disease mortality and its associated factors in selected Asian countries: Evidence from panel error correction model. BMC Public Health 2021, 21, 53. [Google Scholar] [CrossRef] [PubMed]


| Author | Study Design | Study Objective | Participants | Age | Ethnicity Country | Sample Type | Microbial Analysis | Key Findings | Limitations | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Brindisi et al. [8] | Cross-sectional | Influence of passive smoke on the nasal microbial composition in children with AR | AR: 50 (25 PSE vs. 25 negatives) | 6–16 years | Unknown, Italy | Anterior nasal swab | 16S rRNA amplification | Children with a PSE: ↓ biodiversity and shift in the microbiota composition. AR PSE nasal microbiota: abundance of Staphylococcus epidermidis and Serratia quinivorans. AR Non-PSE nasal microbiota: abundance of Moraxella nonliquefaciens. | Small sample size. Monocentric evaluation. No ethnicity records. | Fair |
| Marazzato et al. [12] | Case control | Evaluate the microbial composition in the anterior nares of pediatric subjects suffering from AR, AH or both diseases | AR: 19 AH: 20 Both: 13 HC: 13 | 6–12 years | Unknown, Italy | Anterior nasal swab | 16S rRNA amplification of the V3–V4 region | Children with AR and AH show similar alterations in nasal microbiota. Healthy nasal microbiota: abundance of Moraxella nonliquefaciens and Corynebacterium pseudodiphtericum. AR and AH nasal microbiota: abundance of Acinetobacter guillouiae, A. gerneri and Pseudomonas brenneri. | Small sample size. No ethnicity records. | Good |
| Morin et al. [17] | Prospective cohort | Evaluate the development of AR at age 6 years based on early-life microbial exposures | 1 week: 29 AR vs. 332 HC 1 month: 38 AR vs. 403 HC 3 months: 38 AR vs. 407 HC | 1 week, 1 month, and 3 months | European ancestry, Denmark | Fluid aspirated with a soft catheter passed through the nose to the hypopharyngeal region | 16S rRNA amplification of the V4 region | Early-life nasal microbiome diversity is lower in children who develop AR by age 6 years. 1 week: abundance of Streptococcus and Veillonella. Relationship between DNAm and microbial diversity only at 1 week, but not at the other time points or with other diversity metrics. | V4 region is considered a relatively low informative region for taxonomic assignment. DNAm profiles and microbiome composition measured in different upper airway niches (inferior turbinate vs. hypopharynx, respectively) | Good |
| Ta et al. [18] | Case control | Evaluate the development of the nasal microbiota with AR and wheeze over 7 time points (3 weeks and 3, 6, 9, 12, 15, and 18 months) in the first 18 months | AR with wheeze: 34 AR without wheeze: 28 HC: 60 | 3 weeks and 3, 6, 9, 12, 15, 18 months | Chinese, Malay, and Indian, Singapore | Anterior nasal swab | 16S rRNA amplification of the V3–V6 region | Nasal microbiome diversity HC: ↑ over time. Nasal microbiome diversity both AR groups: ↓over time. Although differed in bacterial composition. AR: ↑ in abundance of Oxalobacteraceae and Aerococcaceae. HC: ↑ in abundance of Corynebacteriaceae and early colonization with the Staphylococcaceae. Nasal microbiome is involved in development of early-onset rhinitis and wheeze in infants. | Difficulties in identification all bacterial groups down to the species level. | Good |
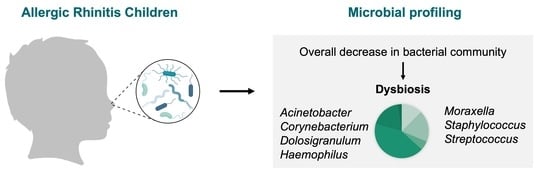
| Yau et al. [14] | Cross-sectional | Evaluate the changes in nasal and ocular surface microbiome with ARC | ARC: 23 HC: 17 | 6–18 years | Unknown, China | Nasopharyngeal nasal swab | 16S rRNA amplification of the V3–V4 region | ARC: Nasal microbiome ∼ ocular microbiome, but ≠ in HC. Most abundant genus: Moraxella (HC 53%; ARC 32%), Corynebacterium (HC 21%; ARC 13%), Dolosigranulum (HC 11%; ARC 11%), Haemophilus (HC 8%; ARC 5%), Streptococcus (HC 3%; ARC 7%), Staphylococcus (HC 2%; ARC 14%) | Small sample size. No ethnicity records. | Good |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azevedo, A.C.; Hilário, S.; Gonçalves, M.F.M. Microbiome in Nasal Mucosa of Children and Adolescents with Allergic Rhinitis: A Systematic Review. Children 2023, 10, 226. https://doi.org/10.3390/children10020226
Azevedo AC, Hilário S, Gonçalves MFM. Microbiome in Nasal Mucosa of Children and Adolescents with Allergic Rhinitis: A Systematic Review. Children. 2023; 10(2):226. https://doi.org/10.3390/children10020226
Chicago/Turabian StyleAzevedo, André Costa, Sandra Hilário, and Micael F. M. Gonçalves. 2023. "Microbiome in Nasal Mucosa of Children and Adolescents with Allergic Rhinitis: A Systematic Review" Children 10, no. 2: 226. https://doi.org/10.3390/children10020226