Heat Shock Cognate 70 kDa Protein Is the Target of Tetradecyl 2,3-Dihydroxybenzoate for Neuritogenic Effect in PC12 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Microarray Analysis
2.3. Preparation of ABG-001 and the Probes
2.4. Evaluation of the Neuritogenic Activity
2.5. In Situ Fluorescence Labeling Using ABG-P1 and ABG-PC
2.6. Pull Down Assay
2.7. Western Blot Analysis
2.8. Cellular Thermal Shift Assay
2.9. RNA Interference
2.10. Statistical Analysis
3. Results
3.1. Microarray Analytical Results after Treatment with ABG-001 in PC12 Cells
3.2. Chemical Synthesis and Biological Evaluation Test of ABG-001 Based Probes in PC12 Cells
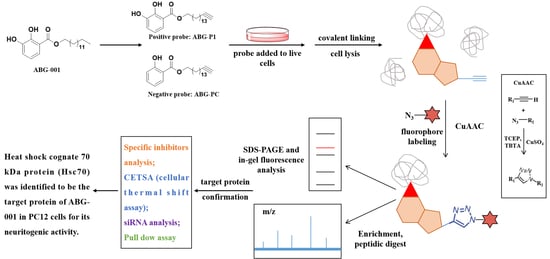
3.3. Target Protein Identification of ABG-001 by ABPP
3.4. Target Confirmation of ABG-001 by CETSA, siRNA and Inhibitor Analysis
3.5. Target Identification of ABG-001 by Pull-Down Assay
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Xu, C.J.; Wang, J.L.; Jin, W.L. The emerging therapeutic role of NGF in Alzheimer’s Disease. Neurochem. Res. 2016, 41, 1211–1218. [Google Scholar] [CrossRef]
- Aloe, L.; Rocco, M.L.; Balzamino, B.O.; Micera, A. Nerve growth factor: A focus on neuroscience and therapy. Curr. Neuropharmacol. 2015, 13, 294–303. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.J.; Li, J.Y.; Qi, J.H. Gentisides A and B, two new neuritogenic compounds from the traditional Chinese medicine Gentiana rigescens Franch. Bioorg. Med. Chem. 2010, 18, 2131–2134. [Google Scholar] [CrossRef]
- Gao, L.J.; Xiang, L.; Luo, Y.; Wang, G.F.; Li, J.Y.; Qi, J.H. Gentisides C-K: Nine new neuritogenic compounds from the traditional Chinese medicine Gentiana rigescens Franch. Bioorg. Med. Chem. 2010, 18, 6995–7000. [Google Scholar] [CrossRef]
- Luo, Y.; Sun, K.Y.; Li, L.; Gao, L.J.; Wang, G.F.; Qu, Y.; Xiang, L.; Chen, L.; Hu, Y.Z.; Qi, J. Structure-activity relationships of neuritogenic Gentiside derivatives. ChemMedChem 2011, 6, 1986–1989. [Google Scholar] [CrossRef]
- Danese, S.; Fiocchi, C.; Panes, J. Drug development in IBD: From novel target identification to early clinical trials. Gut 2016, 65, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.Q.; Gao, L.J.; Kawatani, M.; Chen, J.Z.; Cao, X.L.; Osada, H.; Xiang, L.; Qi, J. Neuritogenic activity of tetradecyl 2,3-dihydroxybenzoate is mediated through the insulin-like growth factor 1 receptor/phosphatidylinositol 3 kinase/mitogen-activated protein kinase signaling pathway. Mol. Pharmacol. 2015, 88, 326–334. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Wang, F.; Tao, S. Proteome microarray technology and application: Higher, wider, and deeper. Expert Rev. Proteomics 2019, 16, 815–827. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-based protein profiling: From enzyme chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonovic, M.; Bogyo, M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteomics 2008, 5, 721–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornoe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Molina, D.M. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.H.; Osada, H.; Xing, T.Y.; Yoshida, M.; Xiang, L.; Qi, J. The insulin receptor: A potential target of amarogentin isolated from Gentiana rigescens Franch that induces neurogenesis in PC12 cells. Biomedicines 2021, 9, 581. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, R.; Scholz, S.R.; Dahmen, H.; Wegener, A.; Sirrenberg, C.; Musil, D.; Bomke, J.; Eggenweiler, H.M.; Mayer, M.P.; Bukau, B. Functional analysis of Hsp70 inhibitors. PLoS ONE 2013, 8, e78443. [Google Scholar] [CrossRef]
- Cerezo, M.; Lehraiki, A.; Millet, A.; Rouaud, F.; Plaisant, M.; Jaune, E.; Botton, T.; Ronco, C.; Abbe, P.; Amdouni, H.; et al. Compounds triggering ER stress exert anti-melanoma effects and overcome BRAF inhibitor resistance. Cancer Cell 2016, 29, 805–819. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gao, L.J.; Sun, K.Y.; Xiao, D.; Li, W.Y.; Xiang, L.; Qi, J.H. Benzoate fraction from Gentiana rigescens Franch alleviates scopolamine induced impaired memory in mice model in vivo. J. Ethnopharmacol. 2016, 193, 107–116. [Google Scholar] [CrossRef]
- Xiang, L.; Li, J.; Wang, Y.H.; Tang, R.Q.; Wang, Q.; Wu, Q.B.; Qi, J.H. Tetradecyl 2,3-dihydroxybenzoate improves the symptoms of diabetic mice by modulation of insulin and adiponectin signaling pathways. Front. Pharmacol. 2017, 8, 806. [Google Scholar] [CrossRef]
- Youssef, M.I.; Zhou, Y.; Eissa, I.H.; Wang, Y.; Zhang, J.; Jiang, L.; Hu, W.; Qi, J.; Chen, Z. Tetradecyl 2,3-dihydroxybenzoate alleviates oligodendrocyte damage following chronic cerebral hypoperfusion through IGF-1 receptor. Neurochem. Int. 2020, 138, 104749. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, S.; Peng, L.; Xie, C.; Gao, L.; Sun, H.; Lin, L.; Ding, K.; Li, Z. Development and application of novel electrophilic warheads in target identification and drug discovery. Biochem. Pharmacol. 2021, 190, 114636. [Google Scholar] [CrossRef]
- Ha, J.; Park, H.; Park, J.; Park, S.B. Recent advances in identifying protein targets in drug discovery. Cell Chem. Biol. 2021, 28, 394–423. [Google Scholar] [CrossRef] [PubMed]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, T.B.; Krueger-Naug, A.M.; Clarke, D.B.; Arrigo, A.P.; Currie, R.W. The role of heat shock proteins Hsp70 and Hsp27 in cellular protection of the central nervous system. Int. J. Hyperth. 2005, 21, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Piedrahita, D.; Castro-Alvarez, J.F.; Boudreau, R.L.; Villegas-Lanau, A.; Kosik, K.S.; Gallego-Gomez, J.C.; Cardona-Gomez, G.P. β-Secretase 1’s targeting reduces hyperphosphorilated tau, implying autophagy actors in 3xTg-AD mice. Front. Cell. Neurosci. 2016, 9, 498. [Google Scholar] [CrossRef] [Green Version]
- Magrane, J.; Smith, R.C.; Walsh, K.; Querfurth, H.W. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed β-amyloid in neurons. J. Neurosci. 2004, 24, 1700–1706. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.C.; Rosen, K.M.; Pola, R.; Magrané, J. Stress proteins in Alzheimer’s disease. Int. J. Hyperth. 2005, 21, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.M.; Dice, J.F. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000, 275, 31505–31513. [Google Scholar] [CrossRef] [Green Version]
- Massey, A.C.; Kaushik, S.; Sovak, G.; Kiffin, R.; Cuervo, A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 2006, 103, 5805–5810. [Google Scholar] [CrossRef] [Green Version]
- Dou, J.; Su, P.; Xu, C.; Wen, Z.; Mao, Z.; Li, W. Targeting Hsc70-based autophagy to eliminate amyloid β oligomers. Biochem. Biophys. Res. Commun. 2020, 524, 923–928. [Google Scholar] [CrossRef]
- Cheng, L.H.; Muroi, M.; Cao, S.N.; Bian, L.L.; Osada, H.; Xiang, L.; Qi, J.H. 3β,23,28-Trihydroxy-12-oleanene 3β-Caffeate from Desmodium sambuense-induced neurogenesis in PC12 cells mediated by ER stress and BDNF-TrkB signaling pathways. Mol. Pharm. 2019, 16, 1423–1432. [Google Scholar] [CrossRef]
- Li, J.; Sun, K.Y.; Muroi, M.; Gao, L.J.; Chang, Y.T.; Osada, H.; Xiang, L.; Qi, J.H. Cucurbitacin B induces neurogenesis in PC12 cells and protects memory in APP/PS1 mice. J. Cell. Mol. Med. 2019, 23, 6283–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Protein Names | Gene Names | MW [kDa] | Protein Score | Sequence Coverage (%) | Unique Peptides | Peptides | PSMs | |
|---|---|---|---|---|---|---|---|---|
| P63018 | Heat shock cognate 71 kDa protein (Hsc70) | Hspa8 | 70.83 | 2904.42 | 37.15 | 20 | 24 | 70 |
| P06761 | Endoplasmic reticulum chaperone BiP (GRP78) | Hspa5 | 72.30 | 2197.30 | 38.07 | 21 | 23 | 48 |
| F1M953 | Stress-70 protein, mitochondrial | Hspa9 | 73.70 | 924.84 | 34.90 | 18 | 18 | 21 |
| G3V8L3 | Lamin A, isoform CRA_b | Lmna | 74.27 | 837.03 | 29.77 | 16 | 16 | 19 |
| Q9EPH8 | Polyadenylate-binding protein 1 | Pabpc1 | 70.66 | 717.99 | 28.30 | 13 | 16 | 17 |
| Protein Names | Gene Names | MW [kDa] | Protein Score | Sequence Coverage (%) | Unique Peptides | Peptides | PSMs | |
|---|---|---|---|---|---|---|---|---|
| P68255 | 14-3-3 protein theta | Ywhaq | 27.76 | 693.16 | 33.47 | 7 | 9 | 15 |
| G3V913 | Heat shock 27 kDa protein 1 | Hspb1 | 22.79 | 495.84 | 36.89 | 7 | 7 | 12 |
| P68511 | 14-3-3 protein eta | Ywhah | 28.19 | 468.72 | 29.67 | 6 | 8 | 11 |
| P63102 | 14-3-3 protein zeta/delta | Ywhaz | 27.75 | 449.39 | 29.8 | 6 | 8 | 13 |
| P35213 | 14-3-3 protein beta/alpha | Ywhab | 28.04 | 439.91 | 29.67 | 4 | 6 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, L.; Wang, Y.; Xiang, L.; Qi, J. Heat Shock Cognate 70 kDa Protein Is the Target of Tetradecyl 2,3-Dihydroxybenzoate for Neuritogenic Effect in PC12 Cells. Biomedicines 2021, 9, 1483. https://doi.org/10.3390/biomedicines9101483
Cheng L, Wang Y, Xiang L, Qi J. Heat Shock Cognate 70 kDa Protein Is the Target of Tetradecyl 2,3-Dihydroxybenzoate for Neuritogenic Effect in PC12 Cells. Biomedicines. 2021; 9(10):1483. https://doi.org/10.3390/biomedicines9101483
Chicago/Turabian StyleCheng, Lihong, Yanhui Wang, Lan Xiang, and Jianhua Qi. 2021. "Heat Shock Cognate 70 kDa Protein Is the Target of Tetradecyl 2,3-Dihydroxybenzoate for Neuritogenic Effect in PC12 Cells" Biomedicines 9, no. 10: 1483. https://doi.org/10.3390/biomedicines9101483