Advances on Liquid Biopsy Analysis for Glioma Diagnosis
Abstract
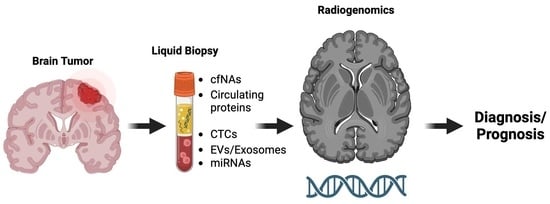
:1. Introduction
2. Basic Principles and Applications of Liquid Biopsy
2.1. Analytes Detected in Biofluids by Liquid Biopsy
2.1.1. Cell-Free DNA (cfDNA), Cell-Free RNA (cfRNA), and Circulating Proteins
2.1.2. Circulating Tumor Cells (CTCs)
2.1.3. Extracellular Vesicles (EVs)/Exosomes and miRNAs
3. Biomarkers Detected through Liquid Biopsy Analysis for the Diagnosis of Gliomas
3.1. cfDNA, cfRNA, and Circulating Proteins in the Diagnosis of Gliomas
3.1.1. cfDNA
3.1.2. cfRNA
3.1.3. Circulating Proteins
3.2. CTCs in Glioma Diagnosis
3.3. Exosomes in Glioma Diagnosis
4. Combination of Radiogenomics and Liquid Biopsy for Glioma Diagnosis
5. Conclusions: Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary Brain Tumours in Adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, Q.; Xu, D.; Li, Y.; Fan, Y.; Li, W.; Yin, X.; Zhang, Y.; Liu, J.; Li, X.; et al. Antitumor Effect of the Newcastle Disease Viral Hemagglutinin-Neuraminidase Gene Is Expressed through an Oncolytic Adenovirus Effect in Osteosarcoma Cells. Anticancer Drugs 2018, 29, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Ammirati, M.; Chotai, S.; Newton, H.; Lamki, T.; Wei, L.; Grecula, J. Hypofractionated Intensity Modulated Radiotherapy with Temozolomide in Newly Diagnosed Glioblastoma Multiforme. J. Clin. Neurosci. 2014, 21, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Mannas, J.P.; Lightner, D.D.; Defrates, S.R.; Pittman, T.; Villano, J.L. Long-Term Treatment with Temozolomide in Malignant Glioma. J. Clin. Neurosci. 2014, 21, 121–123. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and Localization of Surgically Resectable Cancers with a Multi-Analyte Blood Test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-Generation Sequencing in Liquid Biopsy: Cancer Screening and Early Detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef]
- Perakis, S.; Speicher, M.R. Emerging Concepts in Liquid Biopsies. BMC Med. 2017, 15, 75. [Google Scholar] [CrossRef]
- Hirahata, T.; ul Quraish, R.; UI Quraish, A.; UI Quraish, S.; Naz, M.; Razzaq, M.A. Liquid Biopsy: A Distinctive Approach to the Diagnosis and Prognosis of Cancer. Cancer Inf. 2022, 21, 11769351221076062. [Google Scholar] [CrossRef]
- Schilsky, R.L.; Longo, D.L. Closing the Gap in Cancer Genomic Testing. N. Engl. J. Med. 2022, 387, 2107–2110. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Primary Brain Tumors. Cancers 2021, 13, 5429. [Google Scholar] [CrossRef]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid Biopsy: A Step Closer to Transform Diagnosis, Prognosis and Future of Cancer Treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Bauman, M.M.J.; Bouchal, S.M.; Monie, D.D.; Aibaidula, A.; Singh, R.; Parney, I.F. Strategies, Considerations, and Recent Advancements in the Development of Liquid Biopsy for Glioblastoma: A Step towards Individualized Medicine in Glioblastoma. Neurosurg. Focus 2022, 53, E14. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Bast, R.C.; Desch, C.E.; Fritsche, H., Jr.; Kemeny, N.E.; Jessup, J.M.; Locker, G.Y.; Macdonald, J.S.; Mennel, R.G.; Norton, L.; et al. Tumor Marker Utility Grading System: A Framework to Evaluate Clinical Utility of Tumor Markers. JNCI J. Natl. Cancer Inst. 1996, 88, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Febbo, P.G.; Ladanyi, M.; Aldape, K.D.; De Marzo, A.M.; Hammond, M.E.; Hayes, D.F.; Iafrate, A.J.; Kelley, R.K.; Marcucci, G.; Ogino, S.; et al. NCCN Task Force Report: Evaluating the Clinical Utility of Tumor Markers in Oncology. J. Natl. Compr. Cancer Netw. 2011, 9 (Suppl. S5), S1–S32. [Google Scholar] [CrossRef] [PubMed]
- Jelski, W.; Mroczko, B. Molecular and Circulating Biomarkers of Brain Tumors. Int. J. Mol. Sci. 2021, 22, 7039. [Google Scholar] [CrossRef]
- Westphal, M.; Lamszus, K. Circulating Biomarkers for Gliomas. Nat. Rev. Neurol. 2015, 11, 556–566. [Google Scholar] [CrossRef]
- Aarthy, R.; Mani, S.; Velusami, S.; Sundarsingh, S.; Rajkumar, T. Role of Circulating Cell-Free DNA in Cancers. Mol. Diagn. Ther. 2015, 19, 339–350. [Google Scholar] [CrossRef]
- Rykova, E.Y.; Morozkin, E.S.; Ponomaryova, A.A.; Loseva, E.M.; Zaporozhchenko, I.A.; Cherdyntseva, N.V.; Vlassov, V.V.; Laktionov, P.P. Cell-Free and Cell-Bound Circulating Nucleic Acid Complexes: Mechanisms of Generation, Concentration and Content. Expert Opin. Biol. Ther. 2012, 12 (Suppl. S1), S141–S153. [Google Scholar] [CrossRef]
- Roth, C.; Pantel, K.; Müller, V.; Rack, B.; Kasimir-Bauer, S.; Janni, W.; Schwarzenbach, H. Apoptosis-Related Deregulation of Proteolytic Activities and High Serum Levels of Circulating Nucleosomes and DNA in Blood Correlate with Breast Cancer Progression. BMC Cancer 2011, 11, 4. [Google Scholar] [CrossRef]
- Carpenter, E.L.; Bagley, S.J. Clinical Utility of Plasma Cell-Free DNA in Gliomas. Neuro-Oncol. Adv. 2022, 4 (Suppl. S2), ii41–ii44. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The Emerging Role of Cell-Free DNA as a Molecular Marker for Cancer Management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The Diverse Origins of Circulating Cell-Free DNA in the Human Body: A Critical Re-Evaluation of the Literature. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1649–1683. [Google Scholar] [CrossRef] [PubMed]
- Mair, R.; Mouliere, F. Cell-Free DNA Technologies for the Analysis of Brain Cancer. Br. J. Cancer 2022, 126, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, P.; Cheng, S.H.; Cheng, T.H.T.; Wong, J.; Wong, V.W.S.; Ng, S.S.M.; Ma, B.B.Y.; Leung, T.Y.; Chan, S.L.; et al. Orientation-Aware Plasma Cell-Free DNA Fragmentation Analysis in Open Chromatin Regions Informs Tissue of Origin. Genome Res. 2019, 29, 418–427. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced Detection of Circulating Tumor DNA by Fragment Size Analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [PubMed]
- Euskirchen, P.; Bielle, F.; Labreche, K.; Kloosterman, W.P.; Rosenberg, S.; Daniau, M.; Schmitt, C.; Masliah-Planchon, J.; Bourdeaut, F.; Dehais, C.; et al. Same-Day Genomic and Epigenomic Diagnosis of Brain Tumors Using Real-Time Nanopore Sequencing. Acta Neuropathol. 2017, 134, 691–703. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Zhang, M.; McMahon, K.W.; Kinde, I.; Dobbyn, L.; Ptak, J.; Brem, H.; Chaichana, K.; Gallia, G.L.; et al. Detection of Tumor-Derived DNA in Cerebrospinal Fluid of Patients with Primary Tumors of the Brain and Spinal Cord. Proc. Natl. Acad. Sci. USA 2015, 112, 9704–9709. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Oppenheim, F.G. Saliva: A Dynamic Proteome. J. Dent. Res. 2007, 86, 680–693. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Gwak, H.; Lee, J.; Kwak, B.; Jung, H.-I. Salivary Exosome and Cell-Free DNA for Cancer Detection. Micromachines 2018, 9, 340. [Google Scholar] [CrossRef]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and Comparison of in Vitro Degradation Kinetics of DNA in Serum, Urine and Saliva: A Qualitative Study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Nadano, D.; Yasuda, T.; Kishi, K. Measurement of Deoxyribonuclease I Activity in Human Tissues and Body Fluids by a Single Radial Enzyme-Diffusion Method. Clin. Chem. 1993, 39, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Laktionov, P.P. Extracellular Nucleic Acids in Urine: Sources, Structure, Diagnostic Potential. Acta Naturae 2015, 7, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ning, J.; Luo, X.; Du, H.; Zhang, Q.; Zhou, G.; Du, Q.; Ou, Z.; Wang, L.; Wang, Y. New method to preserve the original proportion and integrity of urinary cell-free DNA. J. Clin. Lab. Anal. 2018, 33, e22668. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Lee, E.-J.; Yoon, H.; Lee, D.H.; Kim, K.H. Comparison of Four Commercial Kits for Isolation of Urinary Cell-Free DNA and Sample Storage Conditions. Diagnostics 2020, 10, 234. [Google Scholar] [CrossRef]
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and Circulating Biomarkers in Patients with Glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A Comprehensive Profile of Circulating RNAs in Human Serum. RNA Biol. 2018, 15, 242–250. [Google Scholar] [CrossRef]
- Lu, M. Circular RNA: Functions, Applications and Prospects. ExRNA 2020, 2, 1–7. [Google Scholar] [CrossRef]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long Noncoding RNAs in Cancer Metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, M.; Pös, O.; Márton, É.; Buglyó, G.; Soltész, B.; Keserű, J.; Penyige, A.; Szemes, T.; Nagy, B. Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. Int. J. Mol. Sci. 2020, 21, 6827. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics Characterization of Exosome Cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Obenauf, A.C. Metastatic Colonization by Circulating Tumour Cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Nahed, B.V.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Rothenberg, S.M.; Sequist, L.V.; et al. Brain Tumor Cells in Circulation Are Enriched for Mesenchymal Gene Expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef]
- Perryman, L.; Erler, J.T. Brain Cancer Spreads. Sci. Transl. Med. 2014, 6, 247fs28. [Google Scholar] [CrossRef]
- Pantel, K.; Denève, E.; Nocca, D.; Coffy, A.; Vendrell, J.-P.; Maudelonde, T.; Riethdorf, S.; Alix-Panabières, C. Circulating Epithelial Cells in Patients with Benign Colon Diseases. Clin. Chem. 2012, 58, 936–940. [Google Scholar] [CrossRef]
- Keup, C.; Mach, P.; Aktas, B.; Tewes, M.; Kolberg, H.-C.; Hauch, S.; Sprenger-Haussels, M.; Kimmig, R.; Kasimir-Bauer, S. RNA Profiles of Circulating Tumor Cells and Extracellular Vesicles for Therapy Stratification of Metastatic Breast Cancer Patients. Clin. Chem. 2018, 64, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating Tumor Cell Is a Common Property of Brain Glioma and Promotes the Monitoring System. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef] [PubMed]
- Krol, I.; Castro-Giner, F.; Maurer, M.; Gkountela, S.; Szczerba, B.M.; Scherrer, R.; Coleman, N.; Carreira, S.; Bachmann, F.; Anderson, S.; et al. Detection of Circulating Tumour Cell Clusters in Human Glioblastoma. Br. J. Cancer 2018, 119, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Théry, C. Exosomes: Secreted Vesicles and Intercellular Communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer Cell Exosomes Depend on Cell-Surface Heparan Sulfate Proteoglycans for Their Internalization and Functional Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and Exosomes: Shedding the Confusion between Extracellular Vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging Roles of Exosomes in Normal and Pathological Conditions: New Insights for Diagnosis and Therapeutic Applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef]
- Skouras, P.; Gargalionis, A.N.; Piperi, C. Exosomes as Novel Diagnostic Biomarkers and Therapeutic Tools in Gliomas. Int. J. Mol. Sci. 2023, 24, 10162. [Google Scholar] [CrossRef]
- Romano, E.; Netti, P.A.; Torino, E. Exosomes in Gliomas: Biogenesis, Isolation, and Preliminary Applications in Nanomedicine. Pharmaceuticals 2020, 13, 319. [Google Scholar] [CrossRef]
- André, F.; Schartz, N.E.C.; Chaput, N.; Flament, C.; Raposo, G.; Amigorena, S.; Angevin, E.; Zitvogel, L. Tumor-Derived Exosomes: A New Source of Tumor Rejection Antigens. Vaccine 2002, 20 (Suppl. S4), A28–A31. [Google Scholar] [CrossRef]
- Chaput, N.; Taïeb, J.; Schartz, N.E.C.; André, F.; Angevin, E.; Zitvogel, L. Exosome-Based Immunotherapy. Cancer Immunol. Immunother. 2004, 53, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Sheng, L.; Stewart, T.; Zabetian, C.P.; Zhang, J. New Windows into the Brain: Central Nervous System-Derived Extracellular Vesicles in Blood. Prog. Neurobiol. 2019, 175, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Boisselier, B.; Gállego Pérez-Larraya, J.; Rossetto, M.; Labussière, M.; Ciccarino, P.; Marie, Y.; Delattre, J.-Y.; Sanson, M. Detection of IDH1 Mutation in the Plasma of Patients with Glioma. Neurology 2012, 79, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Lavon, I.; Refael, M.; Zelikovitch, B.; Shalom, E.; Siegal, T. Serum DNA Can Define Tumor-Specific Genetic and Epigenetic Markers in Gliomas of Various Grades. Neuro Oncol. 2010, 12, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Balaña, C.; Ramirez, J.L.; Taron, M.; Roussos, Y.; Ariza, A.; Ballester, R.; Sarries, C.; Mendez, P.; Sanchez, J.J.; Rosell, R. O6-Methyl-Guanine-DNA Methyltransferase Methylation in Serum and Tumor DNA Predicts Response to 1,3-Bis(2-Chloroethyl)-1-Nitrosourea but Not to Temozolamide plus Cisplatin in Glioblastoma Multiforme. Clin. Cancer Res. 2003, 9, 1461–1468. [Google Scholar]
- Mouliere, F.; Mair, R.; Chandrananda, D.; Marass, F.; Smith, C.G.; Su, J.; Morris, J.; Watts, C.; Brindle, K.M.; Rosenfeld, N. Detection of Cell-Free DNA Fragmentation and Copy Number Alterations in Cerebrospinal Fluid from Glioma Patients. EMBO Mol. Med. 2018, 10, e9323. [Google Scholar] [CrossRef]
- Shi, G.; Cui, W.; Benchimol, M.; Liu, Y.-T.; Mattrey, R.F.; Mukthavaram, R.; Kesari, S.; Esener, S.C.; Simberg, D. Isolation of Rare Tumor Cells from Blood Cells with Buoyant Immuno-Microbubbles. PLoS ONE 2013, 8, e58017. [Google Scholar] [CrossRef]
- Lynch, D.; Powter, B.; Po, J.W.; Cooper, A.; Garrett, C.; Koh, E.-S.; Sheridan, M.; van Gelder, J.; Darwish, B.; Mckechnie, S.; et al. Isolation of Circulating Tumor Cells from Glioblastoma Patients by Direct Immunomagnetic Targeting. Appl. Sci. 2020, 10, 3338. [Google Scholar] [CrossRef]
- Macarthur, K.M.; Kao, G.D.; Chandrasekaran, S.; Alonso-Basanta, M.; Chapman, C.; Lustig, R.A.; Wileyto, E.P.; Hahn, S.M.; Dorsey, J.F. Detection of Brain Tumor Cells in the Peripheral Blood by a Telomerase Promoter-Based Assay. Cancer Res. 2014, 74, 2152–2159. [Google Scholar] [CrossRef]
- Olioso, D.; Caccese, M.; Santangelo, A.; Lippi, G.; Zagonel, V.; Cabrini, G.; Lombardi, G.; Dechecchi, M.C. Serum Exosomal MicroRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study. Cancers 2021, 13, 3006. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Lan, F.; Hu, M.; Pan, Q.; Wang, Q.; Wang, J. Downregulation of Serum MicroRNA-205 as a Potential Diagnostic and Prognostic Biomarker for Human Glioma. J. Neurosurg. 2016, 124, 122–128. [Google Scholar] [CrossRef]
- Wang, X.; Cao, Q.; Shi, Y.; Wu, X.; Mi, Y.; Liu, K.; Kan, Q.; Fan, R.; Liu, Z.; Zhang, M. Identification of Low-Dose Radiation-Induced Exosomal Circ-METRN and MiR-4709-3p/GRB14/PDGFRα Pathway as a Key Regulatory Mechanism in Glioblastoma Progression and Radioresistance: Functional Validation and Clinical Theranostic Significance. Int. J. Biol. Sci. 2021, 17, 1061–1078. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.; Nguyen, H.; Drummond, K.; Morokoff, A. Circulating Biomarkers for Glioma: A Review. Neurosurgery 2021, 88, E221–E230. [Google Scholar] [CrossRef]
- Faria, G.; Silva, E.; Da Fonseca, C.; Quirico-Santos, T. Circulating Cell-Free DNA as a Prognostic and Molecular Marker for Patients with Brain Tumors under Perillyl Alcohol-Based Therapy. Int. J. Mol. Sci. 2018, 19, 1610. [Google Scholar] [CrossRef] [PubMed]
- Mathios, D.; Phallen, J. Advances in Molecular Biomarkers and Liquid Biopsy in Gliomas. Neurooncol. Adv. 2022, 4 (Suppl. S2), ii15–ii21. [Google Scholar] [CrossRef]
- Li, D.; Bonner, E.R.; Wierzbicki, K.; Panditharatna, E.; Huang, T.; Lulla, R.; Mueller, S.; Koschmann, C.; Nazarian, J.; Saratsis, A.M. Standardization of the Liquid Biopsy for Pediatric Diffuse Midline Glioma Using DdPCR. Sci. Rep. 2021, 11, 5098. [Google Scholar] [CrossRef]
- Panditharatna, E.; Kilburn, L.B.; Aboian, M.S.; Kambhampati, M.; Gordish-Dressman, H.; Magge, S.N.; Gupta, N.; Myseros, J.S.; Hwang, E.I.; Kline, C.; et al. Clinically Relevant and Minimally Invasive Tumor Surveillance of Pediatric Diffuse Midline Gliomas Using Patient Derived Liquid Biopsy. Clin. Cancer Res. 2018, 24, 5850–5859. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Basdra, E.K.; Papavassiliou, A.G.; Piperi, C. Prominent Role of Histone Modifications in the Regulation of Tumor Metastasis. Int. J. Mol. Sci. 2021, 22, 2778. [Google Scholar] [CrossRef]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef] [PubMed]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Morell, M.; O’Shea, K.S.; Moran, J.V.; Gage, F.H. L1 Retrotransposition in Human Neural Progenitor Cells. Nature 2009, 460, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic Retrotransposition Alters the Genetic Landscape of the Human Brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef]
- Zheng, S.; Houseman, E.A.; Morrison, Z.; Wrensch, M.R.; Patoka, J.S.; Ramos, C.; Haas-Kogan, D.A.; McBride, S.; Marsit, C.J.; Christensen, B.C.; et al. DNA Hypermethylation Profiles Associated with Glioma Subtypes and EZH2 and IGFBP2 MRNA Expression. Neuro Oncol. 2011, 13, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Ohka, F.; Natsume, A.; Motomura, K.; Kishida, Y.; Kondo, Y.; Abe, T.; Nakasu, Y.; Namba, H.; Wakai, K.; Fukui, T.; et al. The Global DNA Methylation Surrogate LINE-1 Methylation Is Correlated with MGMT Promoter Methylation and Is a Better Prognostic Factor for Glioma. PLoS ONE 2011, 6, e23332. [Google Scholar] [CrossRef] [PubMed]
- Sabedot, T.S.; Malta, T.M.; Snyder, J.; Nelson, K.; Wells, M.; deCarvalho, A.C.; Mukherjee, A.; Chitale, D.A.; Mosella, M.S.; Sokolov, A.; et al. A Serum-Based DNA Methylation Assay Provides Accurate Detection of Glioma. Neuro Oncol. 2021, 23, 1494–1508. [Google Scholar] [CrossRef]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and Discrimination of Intracranial Tumors Using Plasma Cell-Free DNA Methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef]
- Martínez-Ricarte, F.; Mayor, R.; Martínez-Sáez, E.; Rubio-Pérez, C.; Pineda, E.; Cordero, E.; Cicuéndez, M.; Poca, M.A.; López-Bigas, N.; Ramon, Y.; et al. Molecular Diagnosis of Diffuse Gliomas through Sequencing of Cell-Free Circulating Tumor DNA from Cerebrospinal Fluid. Clin. Cancer Res. 2018, 24, 2812–2819. [Google Scholar] [CrossRef]
- Muralidharan, K.; Yekula, A.; Small, J.L.; Rosh, Z.S.; Kang, K.M.; Wang, L.; Lau, S.; Zhang, H.; Lee, H.; Bettegowda, C.; et al. TERT Promoter Mutation Analysis for Blood-Based Diagnosis and Monitoring of Gliomas. Clin. Cancer Res. 2021, 27, 169–178. [Google Scholar] [CrossRef]
- Fujioka, Y.; Hata, N.; Akagi, Y.; Kuga, D.; Hatae, R.; Sangatsuda, Y.; Michiwaki, Y.; Amemiya, T.; Takigawa, K.; Funakoshi, Y.; et al. Molecular Diagnosis of Diffuse Glioma Using a Chip-Based Digital PCR System to Analyze IDH, TERT, and H3 Mutations in the Cerebrospinal Fluid. J. Neurooncol. 2021, 152, 47–54. [Google Scholar] [CrossRef]
- Husain, A.; Mishra, S.; Hadi, R.; Sahu, A.; Kumari, S.; Rastogi, M.; Khurana, R.; Shukla, S.; Siddiqui, M.H.; Husain, N. Dynamics of Cell-Free DNA in Predicting Response in Adult Diffuse Glioma on Chemoradiotherapy. Cancer Genet. 2022, 268–269, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Palande, V.; Siegal, T.; Detroja, R.; Gorohovski, A.; Glass, R.; Flueh, C.; Kanner, A.A.; Laviv, Y.; Har-Nof, S.; Levy-Barda, A.; et al. Detection of Gene Mutations and Gene–Gene Fusions in Circulating Cell-Free DNA of Glioblastoma Patients: An Avenue for Clinically Relevant Diagnostic Analysis. Mol. Oncol. 2022, 16, 2098–2114. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, Y.; Han, C.; Wang, X.; She, L.; Zhang, H. MiRNA Microarray Reveals Specific Expression in the Peripheral Blood of Glioblastoma Patients. Int. J. Oncol. 2014, 45, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma Specific MiRNAs as Predictive Biomarkers for Diagnosis and Prognosis of Glioma. J. Exp. Clin. Cancer Res. 2012, 31, 97. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, G.; Feng, D.; Qin, H.; Gong, L.; Zhang, J.; Zhang, Z. MicroRNA-21 Expression Is Associated with Overall Survival in Patients with Glioma. Diagn. Pathol. 2013, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Ilhan-Mutlu, A.; Wagner, L.; Wöhrer, A.; Furtner, J.; Widhalm, G.; Marosi, C.; Preusser, M. Plasma MicroRNA-21 Concentration May Be a Useful Biomarker in Glioblastoma Patients. Cancer Investig. 2012, 30, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Mazor, G.; Levin, L.; Picard, D.; Ahmadov, U.; Carén, H.; Borkhardt, A.; Reifenberger, G.; Leprivier, G.; Remke, M.; Rotblat, B. The LncRNA TP73-AS1 Is Linked to Aggressiveness in Glioblastoma and Promotes Temozolomide Resistance in Glioblastoma Cancer Stem Cells. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Shen, J.; Hodges, T.R.; Song, R.; Gong, Y.; Calin, G.A.; Heimberger, A.B.; Zhao, H. Serum HOTAIR and GAS5 Levels as Predictors of Survival in Patients with Glioblastoma. Mol. Carcinog. 2018, 57, 137–141. [Google Scholar] [CrossRef]
- Yuan, Z.; Yang, Z.; Li, W.; Wu, A.; Su, Z.; Jiang, B. Exosome-Mediated Transfer of Long Noncoding RNA HOTAIR Regulates Temozolomide Resistance by MiR-519a-3p/RRM1 Axis in Glioblastoma. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Naryzhny, S.; Ronzhina, N.; Zorina, E.; Kabachenko, F.; Zavialova, M.; Zgoda, V.; Klopov, N.; Legina, O.; Pantina, R. Evaluation of Haptoglobin and Its Proteoforms as Glioblastoma Markers. Int. J. Mol. Sci. 2021, 22, 6533. [Google Scholar] [CrossRef]
- Gautam, P.; Nair, S.C.; Gupta, M.K.; Sharma, R.; Polisetty, R.V.; Uppin, M.S.; Sundaram, C.; Puligopum, A.K.; Ankathi, P.; Purohit, A.K.; et al. Proteins with altered levels in plasma from glioblastoma patients as revealed by iTRAQ-based quantitative proteomic analysis. PLoS ONE 2012, 7, e46153. [Google Scholar] [CrossRef] [PubMed]
- Bellia, F.; Vecchio, G.; Rizzarelli, E. Carnosinases, their substrates and diseases. Molecules 2014, 19, 2299–2329. [Google Scholar] [CrossRef] [PubMed]
- Gállego Pérez-Larraya, J.; Paris, S.; Idbaih, A.; Dehais, C.; Laigle-Donadey, F.; Navarro, S.; Capelle, L.; Mokhtari, K.; Marie, Y.; Sanson, M.; et al. Diagnostic and Prognostic Value of Preoperative Combined GFAP, IGFBP-2, and YKL-40 Plasma Levels in Patients with Glioblastoma. Cancer 2014, 120, 3972–3980. [Google Scholar] [CrossRef]
- Ochieng, J.; Nangami, G.; Sakwe, A.; Moye, C.; Alvarez, J.; Whalen, D.; Thomas, P.; Lammers, P. Impact of Fetuin-A (AHSG) on Tumor Progression and Type 2 Diabetes. Int. J. Mol. Sci. 2018, 19, 2211. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.; Volnitskiy, A.; Kopylov, A.; Zorina, E.; Kamyshinsky, R.; Bairamukov, V.; Garaeva, L.; Shlikht, A.; Shtam, T. Proteome of Glioblastoma-Derived Exosomes as a Source of Biomarkers. Biomedicines 2020, 8, 216. [Google Scholar] [CrossRef]
- Miyauchi, E.; Furuta, T.; Ohtsuki, S.; Tachikawa, M.; Uchida, Y.; Sabit, H.; Obuchi, W.; Baba, T.; Watanabe, M.; Terasaki, T.; et al. Identification of Blood Biomarkers in Glioblastoma by SWATH Mass Spectrometry and Quantitative Targeted Absolute Proteomics. PLoS ONE 2018, 13, e0193799. [Google Scholar] [CrossRef]
- Qin, G.; Li, X.; Chen, Z.; Liao, G.; Su, Y.; Chen, Y.; Zhang, W. Prognostic Value of YKL-40 in Patients with Glioblastoma: A Systematic Review and Meta-Analysis. Mol. Neurobiol. 2017, 54, 3264–3270. [Google Scholar] [CrossRef]
- Müller, C.; Holtschmidt, J.; Auer, M.; Heitzer, E.; Lamszus, K.; Schulte, A.; Matschke, J.; Langer-Freitag, S.; Gasch, C.; Stoupiec, M.; et al. Hematogenous Dissemination of Glioblastoma Multiforme. Sci. Transl. Med. 2014, 6, 247ra101. [Google Scholar] [CrossRef]
- Sastre, J.; Maestro, M.L.; Puente, J.; Veganzones, S.; Alfonso, R.; Rafael, S.; García-Saenz, J.A.; Vidaurreta, M.; Martín, M.; Arroyo, M.; et al. Circulating Tumor Cells in Colorectal Cancer: Correlation with Clinical and Pathological Variables. Ann. Oncol. 2008, 19, 935–938. [Google Scholar] [CrossRef]
- Kichkailo, A.S.; Narodov, A.A.; Komarova, M.A.; Zamay, T.N.; Zamay, G.S.; Kolovskaya, O.S.; Erakhtin, E.E.; Glazyrin, Y.E.; Veprintsev, D.V.; Moryachkov, R.V.; et al. Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells. Mol. Ther. Nucleic Acids 2023, 32, 267–288. [Google Scholar] [CrossRef]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic Acid Aptamers: Clinical Applications and Promising New Horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2019, 58, 2236–2240. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, J.; Morokoff, A.P.; Luwor, R.B.; Zhu, H.-J.; Kaye, A.H.; Stylli, S.S. The Emergent Role of Exosomes in Glioma. J. Clin. Neurosci. 2017, 35, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive Proteome Profiling of Glioblastoma-Derived Extracellular Vesicles Identifies Markers for More Aggressive Disease. J. Neurooncol. 2017, 131, 233–244. [Google Scholar] [CrossRef] [PubMed]
- H Rashed, M.; Bayraktar, E.; K Helal, G.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef]
- Klekner, Á.; Szivos, L.; Virga, J.; Árkosy, P.; Bognár, L.; Birkó, Z.; Nagy, B. Significance of Liquid Biopsy in Glioblastoma—A Review. J. Biotechnol. 2019, 298, 82–87. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Whitehead, C.A.; Kaye, A.H.; Drummond, K.J.; Widodo, S.S.; Mantamadiotis, T.; Vella, L.J.; Stylli, S.S. Extracellular Vesicles and Their Role in Glioblastoma. Crit. Rev. Clin. Lab. Sci. 2019, 57, 227–252. [Google Scholar] [CrossRef]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A.A. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncology 2021, 26, 865–878. [Google Scholar] [CrossRef]
- Quezada, C.; Torres, Á.; Niechi, I.; Uribe, D.; Contreras-Duarte, S.; Toledo, F.; San Martín, R.; Gutiérrez, J.; Sobrevia, L. Role of Extracellular Vesicles in Glioma Progression. Mol. Asp. Med. 2018, 60, 38–51. [Google Scholar] [CrossRef]
- Hagiwara, K.; Ochiya, T.; Kosaka, N. A Paradigm Shift for Extracellular Vesicles as Small RNA Carriers: From Cellular Waste Elimination to Therapeutic Applications. Drug Deliv. Transl. Res. 2014, 4, 31–37. [Google Scholar] [CrossRef]
- Jaiswal, R.; Sedger, L.M. Intercellular Vesicular Transfer by Exosomes, Microparticles and Oncosomes—Implications for Cancer Biology and Treatments. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Putz, U.; Howitt, J.; Doan, A.; Goh, C.-P.; Low, L.-H.; Silke, J.; Tan, S.-S. The Tumor Suppressor PTEN Is Exported in Exosomes and Has Phosphatase Activity in Recipient Cells. Sci. Signal. 2012, 5, ra70. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, P.-Y.; Li, X.-Y.; Chen, J.-X.; Li, Y.; Zhang, X.-Z.; Zhang, C.-G.; Jiang, T.; Li, W.-B.; Ding, W.; et al. Exosomal Levels of MiRNA-21 from Cerebrospinal Fluids Associated with Poor Prognosis and Tumor Recurrence of Glioma Patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef]
- Ivo D’Urso, P.; Fernando D’Urso, O.; Damiano Gianfreda, C.; Mezzolla, V.; Storelli, C.; Marsigliante, S. MiR-15b and MiR-21 as Circulating Biomarkers for Diagnosis of Glioma. Curr. Genom. 2015, 16, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Phillips, S.; Kaimal, V.; Mao, Y.; Hua, W.; Yang, I.; Fu, C.-C.; Nolan, J.; et al. MiRNA Contents of Cerebrospinal Fluid Extracellular Vesicles in Glioblastoma Patients. J. Neurooncol. 2015, 123, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal Transfer of MiR-151a Enhances Chemosensitivity to Temozolomide in Drug-Resistant Glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of Wild-Type EGFR Amplification and EGFRvIII Mutation in CSF-Derived Extracellular Vesicles of Glioblastoma Patients. Neuro Oncol. 2017, 19, 1494–1502. [Google Scholar] [CrossRef]
- Huang, K.; Fang, C.; Yi, K.; Liu, X.; Qi, H.; Tan, Y.; Zhou, J.; Li, Y.; Liu, M.; Zhang, Y.; et al. The Role of PTRF/Cavin1 as a Biomarker in Both Glioma and Serum Exosomes. Theranostics 2018, 8, 1540–1557. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-Based Analysis of Exosomal MRNA Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef] [PubMed]
- Manterola, L.; Guruceaga, E.; Pérez-Larraya, J.G.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A Small Noncoding RNA Signature Found in Exosomes of GBM Patient Serum as a Diagnostic Tool. Neuro Oncol. 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Qing, Q.; Pan, Q.; Hu, M.; Yu, H.; Yue, X. Serum Exosomal MiR-301a as a Potential Diagnostic and Prognostic Biomarker for Human Glioma. Cell. Oncol. 2018, 41, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Shao, N.; Wang, R.; Deng, D.; Xue, L.; Wang, Q.; Zhang, Y.; Shi, Y.; Xia, X.; Wang, S.; et al. Identification of 9 Serum MicroRNAs as Potential Noninvasive Biomarkers of Human Astrocytoma. Neuro Oncol. 2015, 17, 383–391. [Google Scholar] [CrossRef]
- Tzaridis, T.; Reiners, K.S.; Weller, J.; Bachurski, D.; Schäfer, N.; Schaub, C.; Hallek, M.; Scheffler, B.; Glas, M.; Herrlinger, U.; et al. Analysis of Serum MiRNA in Glioblastoma Patients: CD44-Based Enrichment of Extracellular Vesicles Enhances Specificity for the Prognostic Signature. Int. J. Mol. Sci. 2020, 21, 7211. [Google Scholar] [CrossRef]
- Regazzo, G.; Terrenato, I.; Spagnuolo, M.; Carosi, M.; Cognetti, G.; Cicchillitti, L.; Sperati, F.; Villani, V.; Carapella, C.; Piaggio, G.; et al. A Restricted Signature of Serum MiRNAs Distinguishes Glioblastoma from Lower Grade Gliomas. J. Exp. Clin. Cancer Res. 2016, 35, 124. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Hao, J.; Shi, Z.; Wang, Y.; Han, L.; Yu, S.; You, Y.; Jiang, T.; Wang, J.; et al. High Level of MiR-221/222 Confers Increased Cell Invasion and Poor Prognosis in Glioma. J. Transl. Med. 2012, 10, 119. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes Reflect the Hypoxic Status of Glioma Cells and Mediate Hypoxia-Dependent Activation of Vascular Cells during Tumor Development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.; Pei, Y.; Song, W.; Zhang, S. Preparation of a Novel Raman Probe and Its Application in the Detection of Circulating Tumor Cells and Exosomes. ACS Appl. Mater. Interfaces 2019, 11, 28671–28680. [Google Scholar] [CrossRef]
- Shui, L.; Ren, H.; Yang, X.; Li, J.; Chen, Z.; Yi, C.; Zhu, H.; Shui, P. The Era of Radiogenomics in Precision Medicine: An Emerging Approach to Support Diagnosis, Treatment Decisions, and Prognostication in Oncology. Front. Oncol. 2021, 10, 570465. [Google Scholar] [CrossRef]
- Gore, S.; Chougule, T.; Jagtap, J.; Saini, J.; Ingalhalikar, M. A Review of Radiomics and Deep Predictive Modeling in Glioma Characterization. Acad. Radiol. 2021, 28, 1599–1621. [Google Scholar] [CrossRef] [PubMed]
- Jian, A.; Jang, K.; Manuguerra, M.; Liu, S.; Magnussen, J.; Di Ieva, A. Machine Learning for the Prediction of Molecular Markers in Glioma on Magnetic Resonance Imaging: A Systematic Review and Meta-Analysis. Neurosurgery 2021, 89, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Chi, A.S. Radiogenomics Identifying Important Biological Pathways in Gliomas. Neuro Oncol. 2021, 23, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Matsutani, A.; Udagawa, C.; Matsunaga, Y.; Nakamura, S.; Zembutsu, H. Liquid Biopsy for the Detection of Clinical Biomarkers in Early Breast Cancer: New Insights and Challenges. Pharmacogenomics 2020, 21, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Lafata, K.J.; Corradetti, M.N.; Gao, J.; Jacobs, C.D.; Weng, J.; Chang, Y.; Wang, C.; Hatch, A.; Xanthopoulos, E.; Jones, G.; et al. Radiogenomic Analysis of Locally Advanced Lung Cancer Based on CT Imaging and Intratreatment Changes in Cell-Free DNA. Radiol. Imaging Cancer 2021, 3, e200157. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Qian, Z.; Sun, Z.; Xu, K.; Wang, K.; Fan, X.; Zhang, Z.; Li, S.; Wang, Y.; et al. Genotype Prediction of ATRX Mutation in Lower-Grade Gliomas Using an MRI Radiomics Signature. Eur. Radiol. 2018, 28, 2960–2968. [Google Scholar] [CrossRef]
- Calabrese, E.; Villanueva-Meyer, J.E.; Cha, S. A Fully Automated Artificial Intelligence Method for Non-Invasive, Imaging-Based Identification of Genetic Alterations in Glioblastomas. Sci. Rep. 2020, 10, 11852. [Google Scholar] [CrossRef]
- Li, Q.; Dong, F.; Jiang, B.; Zhang, M. Exploring MRI Characteristics of Brain Diffuse Midline Gliomas With the H3 K27M Mutation Using Radiomics. Front. Oncol. 2021, 11, 646267. [Google Scholar] [CrossRef]
- Tian, H.; Wu, H.; Wu, G.; Xu, G. Noninvasive Prediction of TERT Promoter Mutations in High-Grade Glioma by Radiomics Analysis Based on Multiparameter MRI. BioMed Res. Int. 2020, 2020, 3872314. [Google Scholar] [CrossRef]
- Moon, W.-J.; Choi, J.W.; Roh, H.G.; Lim, S.D.; Koh, Y.-C. Imaging Parameters of High Grade Gliomas in Relation to the MGMT Promoter Methylation Status: The CT, Diffusion Tensor Imaging, and Perfusion MR Imaging. Neuroradiology 2012, 54, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Zlatescu, M.; Sijben, A.; Roldan, G.; Easaw, J.; Forsyth, P.; Parney, I.; Sevick, R.; Yan, E.; Demetrick, D.; et al. The Use of Magnetic Resonance Imaging to Noninvasively Detect Genetic Signatures in Oligodendroglioma. Clin. Cancer Res. 2008, 14, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Jiang, C.; Zhang, Y.; Liu, S.; Liu, D.; Liu, Z.; Chen, W.; Liu, P.; Yang, T.; Lyu, Y.; et al. Thin-Slice Magnetic Resonance Imaging-Based Radiomics Signature Predicts Chromosomal 1p/19q Co-Deletion Status in Grade II and III Gliomas. Front. Neurol. 2020, 11, 551771. [Google Scholar] [CrossRef] [PubMed]
- Casale, R.; Lavrova, E.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Development and External Validation of a Non-Invasive Molecular Status Predictor of Chromosome 1p/19q Co-Deletion Based on MRI Radiomics Analysis of Low Grade Glioma Patients. Eur. J. Radiol. 2021, 139, 109678. [Google Scholar] [CrossRef] [PubMed]
- Soffietti, R.; Bettegowda, C.; Mellinghoff, I.K.; Warren, K.E.; Ahluwalia, M.S.; De Groot, J.F.; Galanis, E.; Gilbert, M.R.; Jaeckle, K.A.; Le Rhun, E.; et al. Liquid Biopsy in Gliomas: A RANO Review and Proposals for Clinical Applications. Neuro Oncol. 2022, 24, 855–871. [Google Scholar] [CrossRef]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D’Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers 2022, 14, 3394. [Google Scholar] [CrossRef]


| LB Sample | Sample Source | Biomarker Isolation Technique | Benefit | Reference |
|---|---|---|---|---|
| cfDNA (ctDNA) | Blood, CSF, Saliva | MS/digital PCR, sequencing, LOH ddPCR, MAF, TAS/WES, NGS | Molecular diagnosis, tumor growth, and therapy response monitoring | [27,28,30,63,64,65,66,67] |
| cfRNA | Blood, CSF | Microarray, qPCR | Tumor growth, therapy response monitoring, and molecular diagnosis | [41,42,43,44,62] |
| Circulating proteins | Blood, Urine, CSF, Saliva | qPCR, NGS, microarray | Diagnosis and prognosis | [16,45,46] |
| CTCs | Blood, CSF | EpCAM Immunomagnetic isolation, FISH, density gradient centrifugation/ telomerase activity | Molecular diagnosis, tumor Growth, and therapy response monitoring | [52,53,68,69,70] |
| EVs | Blood, Saliva | qPCR, NGS, microarray | Molecular diagnosis and disease prognosis | [30,58,71,72,73] |
| Sample | Cargo (Biomarker) | Model | Benefit | Reference |
|---|---|---|---|---|
| CSF/Plasma | miRNA-21 | Patients with glioma (grade I to IV) | Characterization of stable versus progressive disease | [120,125,126] |
| CSF | miR-21/miR-103/miR-24/miR-125 | Patients with GB | Disease diagnosis | [126,127] |
| CSF | miR-151a | Patients with GB who received TMZ treatment | Prediction of treatment response | [128] |
| Blood | EGFR/EGFRvIII | In vitro (μNMR) | Characterization of molecular status | [123,129] |
| Blood | PTRF | In vivo mice xenograft | Disease diagnosis | [130] |
| Blood | MGMT, APNG | Patients with GB (microfluidic chip) | Disease prognosis | [76,131] |
| Serum | miR-320/miR-574 3p/RNU6-1 | Patients with GB | Disease diagnosis | [132] |
| Serum | miR-301a | In vitro | Disease prognosis | [133] |
| Serum | miR-15b-5p, miR-16-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p, miR-106a-5p, miR-130-3p, miR-181b-5p, miR-208a-3p | Astrocytoma (grade II to IV) (TaqMan low-density array) | Disease prognosis | [134,135] |
| Serum | miR-497, miR-125b | Patients with GB | Disease prognosis | [136] |
| Plasma | miR-221/222 | Patients with high-grade gliomas | Disease diagnosis, Prediction of poorer prognosis and response to treatment | [137] |
| Plasma | PDGFR, CAV1, IL-8 | In vivo glioma mice xenograft | Disease diagnosis and prognosis | [138] |
| Blood | miR-100 | Patients with GB | Disease diagnosis | [139] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skouras, P.; Markouli, M.; Kalamatianos, T.; Stranjalis, G.; Korkolopoulou, P.; Piperi, C. Advances on Liquid Biopsy Analysis for Glioma Diagnosis. Biomedicines 2023, 11, 2371. https://doi.org/10.3390/biomedicines11092371
Skouras P, Markouli M, Kalamatianos T, Stranjalis G, Korkolopoulou P, Piperi C. Advances on Liquid Biopsy Analysis for Glioma Diagnosis. Biomedicines. 2023; 11(9):2371. https://doi.org/10.3390/biomedicines11092371
Chicago/Turabian StyleSkouras, Panagiotis, Mariam Markouli, Theodosis Kalamatianos, George Stranjalis, Penelope Korkolopoulou, and Christina Piperi. 2023. "Advances on Liquid Biopsy Analysis for Glioma Diagnosis" Biomedicines 11, no. 9: 2371. https://doi.org/10.3390/biomedicines11092371