Increased Stiffness Downregulates Focal Adhesion Kinase Expression in Pancreatic Cancer Cells Cultured in 3D Self-Assembling Peptide Scaffolds
Abstract
:1. Introduction
2. Materials and Methods
2.1. 2D Cell Culture
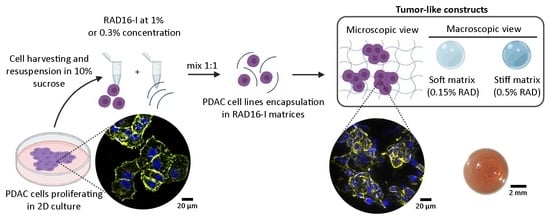
2.2. 3D Cell Culture in the Self-Assembling Peptide Scaffold RAD16-I
2.3. 3D Cell Culture in Collagen Type I
2.4. Protein Binding to RAD16-I Gels
2.5. MTT Assay for Cell Viability and Proliferation
2.6. Immunofluorescence and Image Analysis
2.7. Western Blot
2.8. Statistics
3. Results
3.1. Cell Culture in 3D Self-Assembling Peptide Scaffold RAD16-I
3.2. RAD16-I Scaffold Allows FAK Activation Independent of Matrix Stiffness
3.3. Increased Stiffness Promotes FAK Downregulation in RAD16-I Scaffold
3.4. FAK Expression and Activation Is Maintained Constant Regardless of Stiffness in 3D Collagen Type I Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Makohon-Moore, A.; Iacobuzio-Donahue, C.A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 2016, 16, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Weniger, M.; Honselmann, K.C.; Liss, A.S. The extracellular matrix and pancreatic cancer: A complex relationship. Cancers 2018, 10, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kota, J.; Hancock, J.; Kwon, J.; Korc, M. Pancreatic cancer: Stroma and its current and emerging targeted therapies. Cancer Lett. 2017, 391, 38–49. [Google Scholar] [CrossRef]
- Koikawa, K.; Ohuchida, K.; Ando, Y.; Kibe, S.; Nakayama, H.; Takesue, S.; Endo, S.; Abe, T.; Okumura, T.; Iwamoto, C.; et al. Basement membrane destruction by pancreatic stellate cells leads to local invasion in pancreatic ductal adenocarcinoma. Cancer Lett. 2018, 425, 65–77. [Google Scholar] [CrossRef]
- Zhan, H.X.; Zhou, B.; Cheng, Y.G.; Xu, J.W.; Wang, L.; Zhang, G.Y.; Hu, S.Y. Crosstalk between stromal cells and cancer cells in pancreatic cancer: New insights into stromal biology. Cancer Lett. 2017, 392, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Ohuchida, K.; Mizumoto, K.; Egami, T.; Miyoshi, K.; Moriyama, T.; Cui, L.; Yu, J.; Zhao, M.; Manabe, T.; et al. Tumor-stromal interactions with direct cell contacts enhance proliferation of human pancreatic carcinoma cells. Cancer Sci. 2009, 100, 2309–2317. [Google Scholar] [CrossRef] [PubMed]
- Apte, M.V.; Park, S.; Phillips, P.A.; Santucci, N.; Goldstein, D.; Kumar, R.K.; Ramm, G.A.; Buchler, M.; Friess, H.; McCarroll, J.A.; et al. Desmoplastic Reaction in Pancreatic Cancer: Role of Pancreatic Stellate Cells. Pancreas 2004, 29, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Kikuta, K.; Masamune, A.; Watanabe, T.; Ariga, H.; Itoh, H.; Hamada, S.; Satoh, K.; Egawa, S.; Unno, M.; Shimosegawa, T. Pancreatic stellate cells promote epithelial-mesenchymal transition in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2010, 403, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Pothula, S.P.; Xu, Z.; Goldstein, D.; Pirola, R.C.; Wilson, J.S.; Apte, M.V. Key role of pancreatic stellate cells in pancreatic cancer. Cancer Lett. 2016, 381, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Lachowski, D.; Cortes, E.; Pink, D.; Chronopoulos, A.; Karim, S.A.; Morton, J.P.; Del Río Hernández, A.E. Substrate Rigidity Controls Activation and Durotaxis in Pancreatic Stellate Cells. Sci. Rep. 2017, 7, 2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amrutkar, M.; Aasrum, M.; Verbeke, C.S.; Gladhaug, I.P. Secretion of fibronectin by human pancreatic stellate cells promotes chemoresistance to gemcitabine in pancreatic cancer cells. BMC Cancer 2019, 19, 596. [Google Scholar] [CrossRef] [Green Version]
- Maccurtain, B.M.; Quirke, N.P.; Thorpe, S.D.; Gallagher, T.K. Pancreatic ductal adenocarcinoma: Relating biomechanics and prognosis. J. Clin. Med. 2021, 10, 2711. [Google Scholar] [CrossRef] [PubMed]
- Rubiano, A.; Delitto, D.; Han, S.; Gerber, M.; Galitz, C.; Trevino, J.; Thomas, R.M.; Hughes, S.J.; Simmons, C.S. Viscoelastic properties of human pancreatic tumors and in vitro constructs to mimic mechanical properties. Acta Biomater. 2017, 67, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Takehara, Y.; Kawase, T.; Terashima, K.; Ohkawa, Y.; Hirose, Y.; Koda, A.; Hyodo, N.; Ushio, T.; Hirai, Y.; et al. Feasibility of magnetic resonance elastography for the pancreas at 3T. J. Magn. Reson. Imaging 2016, 43, 384–390. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef] [Green Version]
- Lam, C.R.I.; Wong, H.K.; Nai, S.; Chua, C.K.; Tan, N.S.; Tan, L.P. A 3D biomimetic model of tissue stiffness interface for cancer drug testing. Mol. Pharm. 2014, 11, 2016–2021. [Google Scholar] [CrossRef] [PubMed]
- Guzman, A.; Ziperstein, M.J.; Kaufman, L.J. The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials 2014, 35, 6954–6963. [Google Scholar] [CrossRef]
- Berger, A.J.; Linsmeier, K.M.; Kreeger, P.K.; Masters, K.S. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen. Biomaterials 2017, 141, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.J.; Renner, C.M.; Hale, I.; Yang, X.; Ponik, S.M.; Weisman, P.S.; Masters, K.S.; Kreeger, P.K. Scaffold stiffness influences breast cancer cell invasion via EGFR-linked Mena upregulation and matrix remodeling. Matrix Biol. 2020, 85–86, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Chang, J.K.; Dominguez, A.A.; Lee, H.P.; Nam, S.; Chang, J.; Varma, S.; Qi, L.S.; West, R.B.; Chaudhuri, O. YAP-independent mechanotransduction drives breast cancer progression. Nat. Commun. 2019, 10, 1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.; Pang, E.M.; Adebowale, K.; Wisdom, K.M.; Chaudhuri, O. Increased Stiffness Inhibits Invadopodia Formation and Cell Migration in 3D. Biophys. J. 2020, 119, 726–736. [Google Scholar] [CrossRef]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-king, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrara, B.; Pignatelli, C.; Cossutta, M.; Citro, A.; Courty, J.; Piemonti, L. The extracellular matrix in pancreatic cancer: Description of a complex network and promising therapeutic options. Cancers 2021, 13, 4442. [Google Scholar] [CrossRef]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; Karim, S.A.; Morton, J.P.; Hernández, A.R. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef] [Green Version]
- Gill, B.J.; Gibbons, D.L.; Roudsari, L.C.; Saik, J.E.; Rizvi, Z.H.; Roybal, J.D.; Kurie, J.M.; West, J.L. A synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma model. Cancer Res. 2012, 72, 6013–6023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Tong, X.; Yang, F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using peg-based hydrogels. Mol. Pharm. 2014, 11, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Nguyen, H.D.; Lin, C.C. Dynamic PEG–Peptide Hydrogels via Visible Light and FMN-Induced Tyrosine Dimerization. Adv. Healthc. Mater. 2018, 7, e1800954. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Kang, J.I.; Hwang, B.H.; Park, K.M. Interpenetrating Polymer Network Hydrogels of Gelatin and Poly(ethylene glycol) as an Engineered 3D Tumor Microenvironment. Macromol. Res. 2019, 27, 205–211. [Google Scholar] [CrossRef]
- Puls, T.J.; Tan, X.; Whittington, C.F.; Voytik-Harbin, S.L. 3D collagen fibrillar microstructure guides pancreatic cancer cell phenotype and serves as a critical design parameter for phenotypic models of EMT. PLoS ONE 2017, 12, e0188870. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-K.; Jang, S.D.; Kim, H.; Chung, S.; Park, J.K.; Kuh, H.-J. Phenotypic Heterogeneity and Plasticity of Cancer Cell Migration in a Pancreatic Tumor Three-Dimensional Culture Model. Cancers 2020, 12, 1305. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P.; Hanna, W.; Cramer, G.M.; Celli, J.P. In situ measurement of ECM rheology and microheterogeneity in embedded and overlaid 3D pancreatic tumor stroma co-cultures via passive particle tracking. J. Innov. Opt. Health Sci. 2017, 10, 1742003. [Google Scholar] [CrossRef] [Green Version]
- Carragher, N.O.; Levkau, B.; Ross, R.; Raines, E.W. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp125(FAK), paxillin, and talin. J. Cell Biol. 1999, 147, 619–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieminski, A.L.; Was, A.S.; Kim, G.; Gong, H.; Kamm, R.D. The stiffness of three-dimensional ionic self-assembling peptide gels affects the extent of capillary-like network formation. Cell Biochem. Biophys. 2007, 49, 73–83. [Google Scholar] [CrossRef]
- Genové, E.; Betriu, N.; Semino, C.E. β-Sheet to Random Coil Transition in Self-Assembling Peptide Scaffolds Promotes Proteolytic Degradation. Biomolecules 2022, 12, 411. [Google Scholar] [CrossRef] [PubMed]
- Betriu, N.; Recha-sancho, L.; Semino, C.E. Culturing Mammalian Cells in Three-dimensional Peptide Scaffolds. J. Vis. Exp. 2018, e57259. [Google Scholar] [CrossRef] [PubMed]
- Sieminski, A.L.; Semino, C.E.; Gong, H.; Kamm, R.D. Primary sequence of ionic self-assembling peptide gels affects endothelial cell adhesion and capillary morphogenesis. J. Biomed. Mater. Res.-Part A 2008, 87, 494–504. [Google Scholar] [CrossRef]
- Genové, E.; Schmitmeier, S.; Sala, A.; Borrós, S.; Bader, A.; Griffith, L.G.; Semino, C.E. Functionalized self-assembling peptide hydrogel enhance maintenance of hepatocyte activity in vitro. J. Cell. Mol. Med. 2009, 13, 3387–3397. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Muiños, T.; Recha-Sancho, L.; López-Chicón, P.; Castells-Sala, C.; Mata, A.; Semino, C.E. Bimolecular based heparin and self-assembling hydrogel for tissue engineering applications. Acta Biomater. 2015, 16, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Recha-Sancho, L.; Semino, C.E. Chondroitin sulfate- and decorin-based self-Assembling scaffolds for cartilage tissue engineering. PLoS ONE 2016, 11, e0157603. [Google Scholar] [CrossRef]
- Betriu, N.; Jarrosson-Moral, C.; Semino, C.E. Culture and Differentiation of Human Hair Follicle Dermal Papilla Cells in a Soft 3D Self-Assembling Peptide Scaffold. Biomolecules 2020, 10, 684. [Google Scholar] [CrossRef]
- Castells-Sala, C.; Recha-Sancho, L.; Llucià-Valldeperas, A.; Soler-Botija, C.; Bayes-Genis, A.; Semino, C.E. Three-Dimensional Cultures of Human Subcutaneous Adipose Tissue-Derived Progenitor Cells Based on RAD16-I Self-Assembling Peptide. Tissue Eng. Part C Methods 2016, 22, ten.tec.2015.0270. [Google Scholar] [CrossRef] [PubMed]
- Betriu, N.; Semino, C.E. Development of a 3D Co-Culture System as a Cancer Model Using a Self-Assembling Peptide Scaffold. Gels 2018, 4, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betriu, N.; Andreeva, A.; Semino, C.E. Erlotinib Promotes Ligand-Induced EGFR Degradation in 3D but Not 2D Cultures of Pancreatic Ductal Adenocarcinoma Cells. Cancers 2021, 13, 4504. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manders, E.M.M.; Stap, J.; Brakenhoff, G.J.; Van Driel, R.; Aten, J.A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992, 103, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Sastre, D.; Estadella, I.; Bosch, M.; Felipe, A. Triple-Colocalization Approach to Assess Traffic Patterns and Their Modulation. In Computer Optimized Microscopy, 1st ed.; Rebollo, E., Bosch, M., Eds.; Springer: New York, NY, USA, 2019; pp. 215–233. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Miroshnikova, Y.A.; Jorgens, D.M.; Spirio, L.; Auer, M.; Sarang-Sieminski, A.L.; Weaver, V.M. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys. Biol. 2011, 8, e026013. [Google Scholar] [CrossRef] [Green Version]
- Steele, J.G.; Dalton, B.A.; Johnson, G.; Underwood, P.A. Adsorption of fibronectin and vitronectin onto PrimariaTM and tissue culture polystyrene and relationship to the mechanism of initial attachment of human vein endothelial cells and BHK-21 fibroblasts. Biomaterials 1995, 16, 1057–1067. [Google Scholar] [CrossRef]
- Moreno-Layseca, P.; Streuli, C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014, 34, 144–153. [Google Scholar] [CrossRef]
- Jang, M.; Koh, I.; Lee, J.E.; Lim, J.Y.; Cheong, J.H.; Kim, P. Increased extracellular matrix density disrupts E-cadherin/β-catenin complex in gastric cancer cells. Biomater. Sci. 2018, 6, 2704–2713. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alemany-Ribes, M. Development of 3D Cancer Models to Obtain Mechanistic Insights into Disease Progression. Doctoral Thesis, Ramon Llull University, Barcelona, Spain, 2016. [Google Scholar]
- Jokinen, J.; Dadu, E.; Nykvist, P.; Käpylä, J.; White, D.J.; Ivaska, J.; Vehviläinen, P.; Reunanen, H.; Larjava, H.; Häkkinen, L.; et al. Integrin-mediated cell adhesion to type I collagen fibrils. J. Biol. Chem. 2004, 279, 31956–31963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, S.; Kojima, M.; Higuchi, Y.; Sugimoto, M.; Ikeda, K.; Sakuyama, N.; Takahashi, S.; Hayashi, R.; Ochiai, A.; Saito, N. Assessment of elasticity of colorectal cancer tissue, clinical utility, pathological and phenotypical relevance. Cancer Sci. 2015, 106, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Gordic, S.; Ayache, J.B.; Kennedy, P.; Besa, C.; Wagner, M.; Bane, O.; Ehman, R.L.; Kim, E.; Taouli, B. Value of tumor stiffness measured with MR elastography for assessment of response of hepatocellular carcinoma to locoregional therapy. Abdom. Radiol. 2017, 42, 1685–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leight, J.L.; Wozniak, M.A.; Chen, S.; Lynch, M.L.; Chen, C.S. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol. Biol. Cell 2012, 23, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Fattet, L.; Tsai, J.H.; Guo, Y.; Pai, V.H.; Majeski, H.E.; Chen, A.C.; Sah, R.L.; Taylor, S.S.; Engler, A.J.; et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015, 17, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Nocelo, M.; Raimondo, T.M.; Vining, K.H.; López-López, R.; De La Fuente, M.; Mooney, D.J. Matrix stiffness and tumor-Associated macrophages modulate epithelial to mesenchymal transition of human adenocarcinoma cells. Biofabrication 2018, 10, 035004. [Google Scholar] [CrossRef]
- Markowski, M.C.; Brown, A.C.; Barker, T.H. Directing epithelial to mesenchymal transition through engineered microenvironments displaying orthogonal adhesive and mechanical cues. J. Biomed. Mater. Res.-Part A 2012, 100, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Yangben, Y.; Wang, H.; Zhong, L.; Chiang, M.Y.M.; Tan, Q.; Singh, G.K.; Li, S.; Yang, L. Relative rigidity of cell-substrate effects on hepatic and hepatocellular carcinoma cell migration. J. Biomater. Sci. Polym. Ed. 2012, 24, 148–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaman, M.H.; Trapani, L.M.; Sieminski, A.; MacKellar, D.; Gong, H.; Kamm, R.D.; Wells, A.; Lauffenburger, D.A.; Matsudaira, P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 10889–10894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micalet, A.; Moeendarbary, E.; Cheema, U. 3D In Vitro Models for Investigating the Role of Stiffness in Cancer Invasion. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Jiang, H.; Hegde, S.; Knolhoff, B.L.; Zhu, Y.; Herndon, J.M.; Meyer, M.A.; Nywening, T.M.; Hawkins, W.G.; Shapiro, I.M.; Weaver, D.T.; et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 2016, 22, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Ligorio, C.; Smith, I.P.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels. Gels 2022, 8, 254. [Google Scholar] [CrossRef]
- Grzesiak, J.J.; Bouvet, M. The α2β1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br. J. Cancer 2006, 94, 1311–1319. [Google Scholar] [CrossRef]
- Meltzer, H.; Silberberg, A. Adsorption of collagen, serum albumin, and fibronectin to glass and to each other. J. Colloid Interface Sci. 1988, 126, 292–303. [Google Scholar] [CrossRef]
- Engvall, E.; Ruoslahti, E. Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer 1977, 20, 1–5. [Google Scholar] [CrossRef]
- Velez, D.O.; Ranamukhaarachchi, S.K.; Kumar, A.; Modi, R.N.; Lim, E.W.; Engler, A.J.; Metallo, C.M.; Fraley, S.I. 3D collagen architecture regulates cell adhesion through degradability, thereby controlling metabolic and oxidative stress. Integr. Biol. 2019, 11, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Mi, K.; Wang, G.; Liu, Z.; Feng, Z.; Huang, B.; Zhao, X. Influence of a self-assembling peptide, RADA16, compared with collagen I and matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo. Macromol. Biosci. 2009, 9, 437–443. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betriu, N.; Andreeva, A.; Alonso, A.; Semino, C.E. Increased Stiffness Downregulates Focal Adhesion Kinase Expression in Pancreatic Cancer Cells Cultured in 3D Self-Assembling Peptide Scaffolds. Biomedicines 2022, 10, 1835. https://doi.org/10.3390/biomedicines10081835
Betriu N, Andreeva A, Alonso A, Semino CE. Increased Stiffness Downregulates Focal Adhesion Kinase Expression in Pancreatic Cancer Cells Cultured in 3D Self-Assembling Peptide Scaffolds. Biomedicines. 2022; 10(8):1835. https://doi.org/10.3390/biomedicines10081835
Chicago/Turabian StyleBetriu, Nausika, Anna Andreeva, Anna Alonso, and Carlos E. Semino. 2022. "Increased Stiffness Downregulates Focal Adhesion Kinase Expression in Pancreatic Cancer Cells Cultured in 3D Self-Assembling Peptide Scaffolds" Biomedicines 10, no. 8: 1835. https://doi.org/10.3390/biomedicines10081835