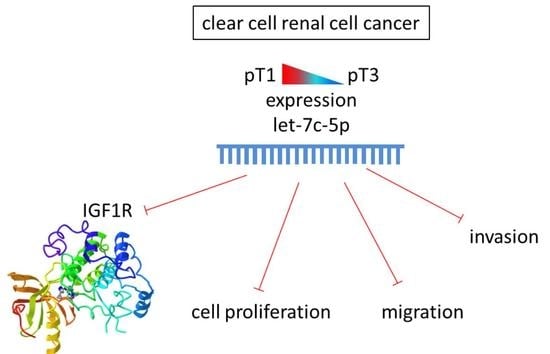
MicroRNA Associated with the Invasive Phenotype in Clear Cell Renal Cell Carcinoma: Let-7c-5p Inhibits Proliferation, Migration, and Invasion by Targeting Insulin-like Growth Factor 1 Receptor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA Isolation and miRNA Screening Analysis of Clinical Samples
2.3. Cell Culture
2.4. Prediction of miRNA Targets
2.5. Cell Transfection of pre-miR Constructs
2.6. Cell Migration and Invasion Assays
2.7. Cell Proliferation Assay
2.8. Western Blot Analysis
2.9. Luciferase Assay
2.10. Data Analysis
3. Results
3.1. Patient Demographics
3.2. miRNA Screening Results
3.3. Transfection of 786-0 and Caki-1 Cells with let-7c-5p Inhibits Proliferation, Migration, and Invasion In Vitro
3.4. IGF1R Expression Is Directly Regulated by let-7c-5p
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cairns, P. Renal Cell Carcinoma. Cancer Biomark. 2011, 9, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Bacik, J.; Mariani, T.; Russo, P.; Mazumdar, M.; Reuter, V. Treatment Outcome and Survival Associated with Metastatic Renal Cell Carcinoma of Non–Clear-Cell Histology. J. Clin. Oncol. 2002, 20, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, J.M.; Miller, D.C.; Daignault, S.; Hollenbeck, B.K. Rising Incidence of Small Renal Masses: A Need to Reassess Treatment Effect. J. Natl. Cancer Inst. 2006, 98, 1331–1334. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grünwald, V.; Gil-lessen, S.; Horwich, A. Renal Cell Carcinoma: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2019, 30, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, W.; Liu, X.; Wang, G.; Fu, B. Impact of Pathological T3a Upstaging on Oncological Outcomes of Clinical T1 Renal Cell Carcinoma: A Meta-Analysis. J. Cancer 2019, 10, 4998–5006. [Google Scholar] [CrossRef]
- Moynihan, M.J.; Sullivan, T.B.; Burks, E.; Schober, J.; Calabrese, M.; Fredrick, A.; Kalantzakos, T.; Warrick, J.; Canes, D.; Raman, J.D.; et al. MicroRNA Profile in Stage I Clear Cell Renal Cell Carcinoma Predicts Progression to Metastatic Disease. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 799.e11–799.e22. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Jung, K. MicroRNAs as Regulators of Signal Transduction in Urological Tumors. Clin. Chem. 2011, 57, 954–968. [Google Scholar] [CrossRef]
- Heinzelmann, J.; Henning, B.; Sanjmyatav, J.; Posorski, N.; Steiner, T.; Wunderlich, H.; Gajda, M.R.; Junker, K. Specific MiRNA Signatures Are Associated with Metastasis and Poor Prognosis in Clear Cell Renal Cell Carcinoma. World J. Urol. 2011, 29, 367–373. [Google Scholar] [CrossRef]
- Slaby, O.; Redova, M.; Poprach, A.; Nekvindova, J.; Iliev, R.; Radova, L.; Lakomy, R.; Svoboda, M.; Vyzula, R. Identification of MicroRNAs Associated with Early Relapse after Nephrectomy in Renal Cell Carcinoma Patients. Genes Chromosom. Cancer 2012, 51, 707–716. [Google Scholar] [CrossRef]
- Peng, J.; Mo, R.; Ma, J.; Fan, J. Let-7b and Let-7c Are Determinants of Intrinsic Chemoresistance in Renal Cell Carcinoma. World J. Surg. Oncol. 2015, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Fedorko, M.; Juracek, J.; Stanik, M.; Svoboda, M.; Poprach, A.; Buchler, T.; Pacik, D.; Dolezel, J.; Slaby, O. Detection of Let-7 MiRNAs in Urine Supernatant as Potential Diagnostic Approach in Non-Metastatic Clear-Cell Renal Cell Carcinoma. Biochem. Med. 2017, 27, 411–417. [Google Scholar] [CrossRef]
- Nadiminty, N.; Tummala, R.; Lou, W.; Zhu, Y.; Shi, X.-B.; Zou, J.X.; Chen, H.; Zhang, J.; Chen, X.; Luo, J.; et al. MicroRNA Let-7c Is Downregulated in Prostate Cancer and Suppresses Prostate Cancer Growth. PLoS ONE 2012, 7, e32832. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Heath, E.; Chen, W.; Cher, M.L.; Powell, I.; Heilbrun, L.; Li, Y.; Ali, S.; Sethi, S.; Hassan, O.; et al. Loss of Let-7 Up-Regulates EZH2 in Prostate Cancer Consistent with the Acquisition of Cancer Stem Cell Signatures That Are Attenuated by BR-DIM. PLoS ONE 2012, 7, e33729. [Google Scholar] [CrossRef] [PubMed]
- Careccia, S.; Mainardi, S.; Pelosi, A.; Gurtner, A.; Diverio, D.; Riccioni, R.; Testa, U.; Pelosi, E.; Piaggio, G.; Sacchi, A.; et al. A Restricted Signature of MiRNAs Distinguishes APL Blasts from Normal Promyelocytes. Oncogene 2009, 28, 4034–4040. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Hou, Y.; Wang, K.; Li, J.; Xia, Y.; Yang, X.; Lv, G.; Xing, X.-L.; Shen, F. Reexpression of Let-7g MicroRNA Inhibits the Proliferation and Migration via K-Ras/HMGA2/Snail Axis in Hepatocellular Carcinoma. BioMed Res. Int. 2014, 2014, 742417. [Google Scholar] [CrossRef]
- Zhao, B.; Han, H.; Chen, J.; Zhang, Z.; Li, S.; Fang, F.; Zheng, Q.; Ma, Y.; Zhang, J.; Wu, N.; et al. MicroRNA Let-7c Inhibits Migration and Invasion of Human Non-Small Cell Lung Cancer by Targeting ITGB3 and MAP4K3. Cancer Lett. 2014, 342, 43–51. [Google Scholar] [CrossRef]
- Jayaraman, M.; Radhakrishnan, R.; Mathews, C.A.; Yan, M.; Husain, S.; Moxley, K.M.; Song, Y.S.; Dhanasekaran, D.N. Identification of Novel Diagnostic and Prognostic MiRNA Signatures in Endometrial Cancer. Genes Cancer 2017, 8, 566–576. [Google Scholar] [CrossRef]
- Liep, J.; Kilic, E.; Meyer, H.A.; Busch, J.; Jung, K.; Rabien, A. Cooperative Effect of MiR-141-3p and MiR-145-5p in the Regulation of Targets in Clear Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0157801. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Hatzigeorgiou, A.G. Online Resources for MiRNA Analysis. Clin. Biochem. 2013, 46, 879–900. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Vejnar, C.E.; Zdobnov, E.M. MiRmap: Comprehensive Prediction of MicroRNA Target Repression Strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of Functional MicroRNA Targets by Integrative Modeling of MicroRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Jeggari, A.; Marks, D.S.; Larsson, E. MiRcode: A Map of Putative MicroRNA Target Sites in the Long Non-Coding Transcriptome. Bioinformatics 2012, 28, 2062–2063. [Google Scholar] [CrossRef]
- Tracz, A.F.; Szczylik, C.; Porta, C.; Czarnecka, A.M. Insulin-like Growth Factor-1 Signaling in Renal Cell Carcinoma. BMC Cancer 2016, 16, 453. [Google Scholar] [CrossRef]
- Hou, B.; Ishinaga, H.; Midorikawa, K.; Nakamura, S.; Hiraku, Y.; Oikawa, S.; Ma, N.; Takeuchi, K.; Murata, M. Let-7c Inhibits Migration and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma by Targeting IGF1R and HMGA2. Oncotarget 2018, 9, 8927–8940. [Google Scholar] [CrossRef]
- Liu, G.-X.; Ma, S.; Li, Y.; Yu, Y.; Zhou, Y.-X.; Lu, Y.-D.; Jin, L.; Wang, Z.-L.; Yu, J.-H. Hsa-Let-7c Controls the Committed Differentiation of IGF-1-Treated Mesenchymal Stem Cells Derived from Dental Pulps by Targeting IGF-1R via the MAPK Pathways. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Liu, G.; Jin, L.; Pang, X.; Wang, Y.; Wang, Z.; Yu, Y.; Yu, J. IGF-1/IGF-1R/Hsa-Let-7c Axis Regulates the Committed Differentiation of Stem Cells from Apical Papilla. Sci. Rep. 2016, 6, 36922. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Cui, L.; Feng, J.; Fan, Q. MicroRNA-182 Suppresses Clear Cell Renal Cell Carcinoma Migration and Invasion by Targeting IGF1R. Neoplasma 2016, 63, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Deng, T.; Li, X.; Cai, W. MiR-193b Inhibits the Growth and Metastasis of Renal Cell Carcinoma by Targeting IGF1R. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2058–2064. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.S.; Cheville, J.C.; Lohse, C.; Cerhan, J.R.; Blute, M.L. Expression of Insulin-like Growth Factor I Receptor and Survival in Patients with Clear Cell Renal Cell Carcinoma. J. Urol. 2003, 170, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Keehn, C.A.; Coppola, D. The Expression of Insulin-like Growth Factor-I Receptor Correlates with Fuhrman Grading of Renal Cell Carcinomas. Hum. Pathol. 2004, 35, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Taliaferro-Smith, L.; Oberlick, E.; Liu, T.; McGlothen, T.; Alcaide, T.; Tobin, R.; Donnelly, S.; Commander, R.; Kline, E.; Nagaraju, G.P.; et al. FAK Activation Is Required for IGF1R-Mediated Regulation of EMT, Migration, and Invasion in Mesenchymal Triple Negative Breast Cancer Cells. Oncotarget 2015, 6, 4757–4772. [Google Scholar] [CrossRef] [PubMed]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the Right Cell Line for Renal Cell Cancer Research. Mol. Cancer 2016, 15, 83. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Lim, S.O.; Yamaguchi, H. Oncogenic signaling pathways associated with immune evasion and resistance to immune checkpoint inhibitors in cancer. Semin. Cancer Biol. 2020, 65, 51–64. [Google Scholar] [CrossRef]
- Peng, X.-S.; Yang, J.-P.; Qiang, Y.-Y.; Sun, R.; Cao, Y.; Zheng, L.-S.; Peng, L.-X.; Lang, Y.-H.; Mei, Y.; Li, C.-Z.; et al. PTPN3 Inhibits the Growth and Metastasis of Clear Cell Renal Cell Carcinoma via Inhibition of PI3K/AKT Signaling. Mol. Cancer Res. 2020, 18, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, G.; Xu, Y.; Li, J.; Ruan, N.; Chen, Y.; Zhang, Q.; Xia, Q. Porcupine Inhibitor LGK974 Downregulates the Wnt Signaling Pathway and Inhibits Clear Cell Renal Cell Carcinoma. BioMed Res. Int. 2020, 2020, 2527643. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, B.; Blystone, S.D.; McHugh, K.P.; Ross, F.P.; Ramos, D.M. Differential expression of alphav integrins in K1735 melanoma cells. Invasion Metastasis 1998, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ivaska, J.; Heino, J. Interplay between cell adhesion and growth factor receptors: From the plasma membrane to the endosomes. Cell Tissue Res. 2010, 339, 111–120. [Google Scholar] [CrossRef]
- Girnita, L.; Worrall, C.; Takahashi, S.; Seregard, S.; Girnita, A. Something old, something new and something borrowed: Emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell. Mol. Life Sci. 2014, 71, 2403–2427. [Google Scholar] [CrossRef]
- Breuksch, I.; Prosinger, F.; Baehr, F.; Engelhardt, F.P.; Bauer, H.K.; Thüroff, J.W.; Heimes, A.S.; Hasenburg, A.; Prawitt, D.; Brenner, W. Integrin α5 triggers the metastatic potential in renal cell carcinoma. Oncotarget 2017, 8, 107530–107542. [Google Scholar] [CrossRef]
- Cox, O.T.; O’Shea, S.; Tresse, E.; Bustamante-Garrido, M.; Kiran-Deevi, R.; O’Connor, R. IGF-1 Receptor and Adhesion Signaling: An Important Axis in Determining Cancer Cell Phenotype and Therapy Resistance. Front. Endocrinol. 2015, 6, 106. [Google Scholar] [CrossRef]
- Zou, X.; Feng, B.; Dong, T.; Yan, G.; Tan, B.; Shen, H.; Huang, A.; Zhang, X.; Zhang, M.; Yang, P.; et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J. Proteom. 2013, 94, 473–485. [Google Scholar] [CrossRef]
- Penet, M.F.; Kakkad, S.; Pathak, A.P.; Krishnamachary, B.; Mironchik, Y.; Raman, V.; Solaiyappan, M.; Bhujwalla, Z.M. Structure and function of a prostate cancer dissemination-permissive extracellular matrix. Clin. Cancer Res. 2017, 23, 2245–2254. [Google Scholar] [CrossRef]
- Xiong, G.; Deng, L.; Zhu, J.; Rychahou, P.G.; Xu, R. Prolyl-4-hydroxylase alpha subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Liu, D.; Liu, Z.; Li, S.; Ge, Y.; Sun, W.; Liu, B. The inhibitory effects of COL1A2 on colorectal cancer cell proliferation, migration, and invasion. J. Cancer 2018, 9, 2953–2962. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Zhao, W.; Shi, B.; Zhang, Z.; Yu, X.; Xie, F.; Guo, Z.; Zhang, X.; Liu, J.; Shen, Q.; et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol. Cancer 2014, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Chang, K.; Ma, J.; Qu, Y.; Xie, H.; Dai, B.; Gan, H.; Zhang, H.; Shi, G.; Zhu, Y.; et al. The Oncogenic Role of COL23A1 in Clear Cell Renal Cell Carcinoma. Sci. Rep. 2017, 7, 9846. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Takada, Y.K.; Fujita, M. Crosstalk between insulin-like growth factor (IGF) receptor and integrins through direct integrin binding to IGF1. Cytokine Growth Factor Rev. 2017, 34, 67–72. [Google Scholar] [CrossRef]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef]
- Tang, Z.; Ow, G.S.; Thiery, J.P.; Ivshina, A.V.; Kuznetsov, V.A. Meta-analysis of Transcriptome Reveals Let-7b as an Unfavorable Prognostic Biomarker and Predicts Molecular and Clinical Sub-Classes in High-Grade Serous Ovarian Carcinoma. Int. J. Cancer 2014, 134, 306–318. [Google Scholar] [CrossRef]
- Hilly, O.; Pillar, N.; Stern, S.; Strenov, Y.; Bachar, G.; Shomron, N.; Shpitzer, T. Distinctive Pattern of Let-7 Family MicroRNAs in Aggressive Carcinoma of the Oral Tongue in Young Patients. Oncol. Lett. 2016, 12, 1729–1736. [Google Scholar] [CrossRef]
- Yuen, J.S.P.; Akkaya, E.; Wang, Y.; Takiguchi, M.; Peak, S.; Sullivan, M.; Protheroe, A.S.; Macau-lay, V.M. Validation of the Type 1 Insulin-like Growth Factor Receptor as a Therapeutic Target in Renal Cancer. Mol. Cancer Ther. 2009, 8, 1448–1459. [Google Scholar] [CrossRef]
- Aleksic, T.; Chitnis, M.M.; Perestenko, O.V.; Gao, S.; Thomas, P.H.; Turner, G.D.; Protheroe, A.S.; Howarth, M.; Macaulay, V.M. Type 1 Insulin-like Growth Factor Receptor Translocates to the Nucleus of Human Tumor Cells. Cancer Res 2010, 70, 6412–6419. [Google Scholar] [CrossRef]
- Yuen, J.S.P.; Cockman, M.E.; Sullivan, M.; Protheroe, A.; Turner, G.D.H.; Roberts, I.S.; Pugh, C.W.; Werner, H.; Macaulay, V.M. The VHL Tumor Suppressor Inhibits Expression of the IGF1R and Its Loss Induces IGF1R Upregulation in Human Clear Cell Renal Carcinoma. Oncogene 2007, 26, 6499–6508. [Google Scholar] [CrossRef] [Green Version]





| Variable | pT1 | pT3 | p-Value |
|---|---|---|---|
| Number of samples | 10 | 10 | |
| Age at surgery, years, mean (range) | 63 (43–76) | 63 (43–78) | 0.912 |
| Gender, n (%) | 0.628 | ||
| Female | 4 (40) | 2 (20) | |
| Male | 6 (60) | 8 (80) | |
| Grade, n (%) | 1 | ||
| 2 | 4 (40) | 3 (30) | |
| 3 | 4 (40) | 4 (40) | |
| 4 | 2 (20) | 3 (30) | |
| tumor size, cm, median (range) | 3.7 (2–5) | 4.1 (2–5) | 0.373 |
| microRNA | FC | p-Value | q-Value |
|---|---|---|---|
| hsa-miR-26a-5p | −1.9 | <0.001 | 0.005 |
| hsa-miR-26b-5p | −1.82 | <0.001 | 0.024 |
| hsa-miR-24-3p | −1.42 | 0.001 | 0.024 |
| hsa-miR-25-3p | 1.84 | 0.001 | 0.024 |
| hsa-let-7a-5p | −1.65 | 0.001 | 0.024 |
| hsa-miR-148b-3p | −1.61 | 0.001 | 0.024 |
| hsa-miR-93-5p | 1.32 | 0.001 | 0.024 |
| hsa-let-7e-5p | −1.58 | 0.002 | 0.038 |
| hsa-let-7c-5p | −1.58 | 0.002 | 0.039 |
| hsa-miR-27a-3p | −1.5 | 0.003 | 0.048 |
| hsa-miR-92a-3p | 1.5 | 0.004 | 0.053 |
| hsa-miR-23b-3p | −1.5 | 0.013 | 0.154 |
| hsa-let-7b-5p | −1.47 | 0.015 | 0.154 |
| hsa-miR-151a-3p | 1.49 | 0.018 | 0.166 |
| hsa-let-7d-5p | −1.31 | 0.022 | 0.188 |
| hsa-miR-106b-5p | 1.27 | 0.025 | 0.201 |
| hsa-miR-484 | 1.65 | 0.033 | 0.227 |
| hsa-miR-101-3p | −1.53 | 0.033 | 0.227 |
| hsa-miR-455-3p | −1.7 | 0.036 | 0.235 |
| hsa-miR-27b-3p | −1.45 | 0.038 | 0.242 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalantzakos, T.J.; Sebel, L.E.; Trussler, J.; Sullivan, T.B.; Burks, E.J.; Sarita-Reyes, C.D.; Canes, D.; Moinzadeh, A.; Rieger-Christ, K.M. MicroRNA Associated with the Invasive Phenotype in Clear Cell Renal Cell Carcinoma: Let-7c-5p Inhibits Proliferation, Migration, and Invasion by Targeting Insulin-like Growth Factor 1 Receptor. Biomedicines 2022, 10, 2425. https://doi.org/10.3390/biomedicines10102425
Kalantzakos TJ, Sebel LE, Trussler J, Sullivan TB, Burks EJ, Sarita-Reyes CD, Canes D, Moinzadeh A, Rieger-Christ KM. MicroRNA Associated with the Invasive Phenotype in Clear Cell Renal Cell Carcinoma: Let-7c-5p Inhibits Proliferation, Migration, and Invasion by Targeting Insulin-like Growth Factor 1 Receptor. Biomedicines. 2022; 10(10):2425. https://doi.org/10.3390/biomedicines10102425
Chicago/Turabian StyleKalantzakos, Thomas J., Luke E. Sebel, James Trussler, Travis B. Sullivan, Eric J. Burks, Carmen D. Sarita-Reyes, David Canes, Alireza Moinzadeh, and Kimberly M. Rieger-Christ. 2022. "MicroRNA Associated with the Invasive Phenotype in Clear Cell Renal Cell Carcinoma: Let-7c-5p Inhibits Proliferation, Migration, and Invasion by Targeting Insulin-like Growth Factor 1 Receptor" Biomedicines 10, no. 10: 2425. https://doi.org/10.3390/biomedicines10102425