Discovery of a Novel Induced Polymorphism in SD1 Gene Governing Semi-Dwarfism in Rice and Development of a Functional Marker for Marker-Assisted Selection
Abstract
:1. Introduction
2. Results
2.1. Monogenic Inheritance of Plant Height
2.2. GA3 Response at the Seedling Stage
2.3. Delineating the Involvement of SD1 Locus
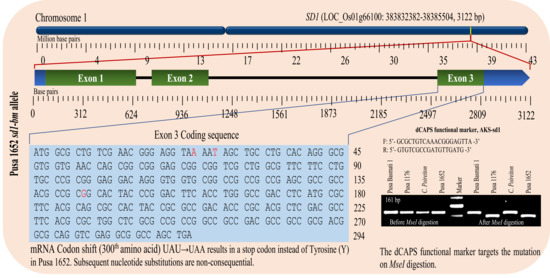
2.4. SD1 Gene Sequence Analysis
2.5. dCAPS Marker Validation of Causal SNP
2.6. Agronomic Effect of sd1-bm Allele
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Analysis of Inheritance of Plant Height
4.3. GA3 Response in the Seedling Stage
4.4. Molecular Analysis and Mapping of the Semi-Dwarfing Gene
4.5. Identification of the Functional SNP in Pusa 1652
4.6. Development and Validation of dCAPS Markers Based on Causal SNP in sd1-bm
4.7. Agronomic Effect of sd1-bm Allele
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, F.; Wang, P.; Zhang, X.; Li, X.; Yan, X.; Fu, D.; Wu, G. The genetic and molecular basis of crop height based on a rice model. Planta 2018, 247, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Ferrero-Serrano, Á.; Cantos, C.; Assmann, S.M. The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb. Perspect. Biol. 2019, 11, a034645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Tang, D.; Liu, K.; Miao, C.; Zhuo, X.; Li, Y.; Tan, X.; Sun, M.; Luo, Q.; Cheng, Z. Characterization of a new semi-dominant dwarf allele of SLR1 and its potential application in hybrid rice breeding. J. Exp. Bot. 2018, 69, 4703–4713. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004, 34, 1642–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashikari, M.; Sakamoto, T. Rice yielding and plant hormones. In Rice Biology in the Genomics Era. Biotechnology in Agriculture and Forestry; Hirano, H.Y., Sano, Y., Hirai, A., Sasaki, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 62, pp. 309–320. [Google Scholar] [CrossRef]
- Castorina, G.; Consonni, G. The Role of Brassinosteroids in Controlling Plant Height in Poaceae: A Genetic Perspective. Int. J. Mol. Sci. 2020, 21, 1191. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shang, L.; Yu, H.; Zeng, L.; Hu, J.; Ni, S.; Rao, Y.; Li, S.; Chu, J.; Meng, X.; et al. A Strigolactone Biosynthesis Gene Contributed to the Green Revolution in Rice. Mol. Plant 2020, 13, 923–932. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semi-dwarf sd-1, “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [Green Version]
- Monna, L.; Kitazawa, N.; Yoshino, R.; Suzuki, J.; Masuda, H.; Maehara, Y.; Tanji, M.; Sato, M.; Nasu, S.; Minobe, Y. Positional Cloning of Rice Semi dwarfing Gene, sd-1: Rice “Green Revolution Gene” Encodes a Mutant Enzyme Involved in Gibberellin Synthesis. DNA Res. 2002, 9, 1117. [Google Scholar] [CrossRef] [Green Version]
- Chandler, R., Jr. The impact of the improved tropical plant type on rice yields in South and Southeast Asia. In Rice Breeding; International Rice Research Institute: Los Banos, Philippines, 1972; pp. 77–85. [Google Scholar]
- Tomita, M.; Ishimoto, K. Rice novel semidwarfing gene d60 can be as effective as green revolution gene sd1. Plants 2019, 8, 464. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zheng, S.; Gui, J.; Fu, C.; Yu, H.; Song, D.; Shen, J.; Qin, P.; Liu, X.; Han, B.; et al. Shortened basal internodes encodes a Gibberellin 2-Oxidase and contributes to lodging resistance in rice. Mol. Plant 2018, 11, 288–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, K.; Takashi, T.; Miura, K.; Qian, Q.; Kitano, H.; Matsuoka, M.; Ashikari, M. Genetic and molecular analysis of the utility of sd1 alleles in rice breeding. Breed. Sci. 2007, 57, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Navasero, S.A.; Garcia, C.V.; Garcia, F.T.; Ramirez, E. Growth habit of the plant in the tropics and its effect on nitrogen response. Tech. Bull. IRRI 1964, 3, 1–80. [Google Scholar]
- Murai, I.M.; Nagano, H.; Onishi, K.; Ogino, A.; Ichikawa, N.; Kc, H.B.; Sano, Y. Differentiation in wild-type allele of the sd1 locus concerning culm length between indica and japonica subspecies of Oryza sativa (rice). Hereditas 2010, 148, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Baghel, S.S. Aromatic rices of Manipur. In A Treatise on the Scented Rices of India; Singh, R.K., Singh, U.S., Eds.; Kalyani Publishers: New Delhi, India, 2003; pp. 347–354. [Google Scholar]
- Choudhury, B.; Khan, M.L.; Dayanandan, S. Genetic structure and diversity of indigenous rice (Oryza sativa) varieties in the Eastern Himalayan region of Northeast India. Springerplus 2013, 2, 228. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, D.; Ray, A. Population genetics analyses of North-East Indian indigenous rice landraces revealed divergent history and alternate origin of aroma in aus group. Plant Genet. Resourc. 2019, 17, 437–447. [Google Scholar] [CrossRef]
- Roy, S.; Marndi, B.C.; Mawkhlieng, B.; Banerjee, A.; Yadav, R.M.; Misra, A.K.; Bansal, K.C. Genetic diversity and structure in hill rice (Oryza sativa L.) landraces from the North-Eastern Himalayas of India. BMC Genet. 2016, 17, 107. [Google Scholar] [CrossRef] [Green Version]
- Ashikari, M.; Sasaki, A.; Tanaka, M.U.; Itoh, H.; Nishimura, A.; Datta, S.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Loss-of-function of a rice Gibberellin Biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘Green revolution’. Breed. Sci. 2002, 52, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Naito, Y.; Tomita, M. Identification of an Isogenic Semi-dwarf Rice Cultivar Carrying the Green Revolution sd1 Gene by Multiplex Codominant ASP-PCR and SSR Markers. Biochem. Genet. 2013, 51, 530–542. [Google Scholar] [CrossRef]
- Wei, L.; Xu, J.; Li, X.; Qian, Q.; Zhu, L. Genetic analysis and mapping of the dominant dwarfing gene D-53 in rice. J. Integr. Plant. Biol. 2006, 48, 447–452. [Google Scholar] [CrossRef]
- Miura, K.; Wu, J.; Sunohara, H.; Wu, X.; Matsumoto, T.; Matsuoka, M.; Ashikari, M.; Kitano, H. High-resolution mapping revealed a 1.3-Mbp genomic inversion in Ssi1, a dominant semidwarf gene in rice (Oryza sativa). Plant Breed. 2009, 128, 63–69. [Google Scholar] [CrossRef]
- Liu, B.M.; Wu, Y.J.; Fu, X.D.; Qian, Q. Characterizations and molecular mapping of a novel dominant semi-dwarf gene Sdd(t) in rice (Oryza sativa). Plant Breed. 2008, 127, 125–130. [Google Scholar] [CrossRef]
- Sunohara, H.; Kawai, T.; Shimizu-Sato, S.; Sato, Y.; Sato, K.; Kitano, H. Dominant mutation of TWISTED DWARF1 encoding an alpha-tubulin protein causes severe dwarfing and right helical growth in rice. Genes Genet. Syst. 2009, 84, 209–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, F.; Xin, X.; Hu, Z.; Xu, J.; Wei, G.; Qian, X.; Yang, J.; He, H.; Luo, X. Genetic analysis and fine mapping of a novel semidominant dwarfing gene LB4D in rice. J. Integr. Plant Biol. 2011, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Asano, K.; Hirano, K.; Ueguchi-Tanaka, M.; Angeles-Shim, R.B.; Komura, T.; Satoh, H.; Kitano, H.; Matusuoka, M.; Ashikari, M. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genom. 2009, 281, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Piao, R.; Chu, S.-H.; Jiang, W.; Yu, Y.; Jin, Y.; Woo, M.O.; Lee, J.; Kim, S.; Koh, H.J. Isolation and Characterization of a Dominant Dwarf Gene, D.-h, in Rice. PLoS ONE 2014, 9, e86210. [Google Scholar] [CrossRef] [PubMed]
- Hayashi-Tsugane, M.; Maekawa, M.; Tsugane, K. A gain-of-function Bushy dwarf tiller 1 mutation in rice microRNA gene miR156d caused by insertion of the DNA transposon nDart1. Sci. Rep. 2015, 5, 14357. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xu, P.; Zhou, J.; Tao, D.; Yu, D. Mapping and breeding value evaluation of a semi-dominant semi-dwarf gene in upland rice. Plant Divers. 2018, 40, 238–244. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, Z.; Chen, L.; Zhou, F.; Zhong, Z.; Jiang, L.; Wan, J. Genetic analysis and fine mapping of a semi-dwarf gene in a centromeric region in rice (Oryza sativa L.). Breed. Sci. 2013, 63, 164–168. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Kumar, P.; Shamim, M.; Tiwari, K.K.; Fatima, P.; Srivastava, D.; Singh, R.; Yadav, P. Genetic diversity and population structure analysis of Asian and African aromatic rice (Oryza sativa L.) genotypes. J. Genet. 2019, 98, 92. [Google Scholar] [CrossRef]
- Singh, V.P.; Satya, P.; Gopala Krishnan, S.; Singh, A.K. Role of Indian Agricultural Research Institute in genetic improvement of rice varieties in India. In Genetic Improvement of Rice Varieties in India; Sharma, S.D., Rao, U.P., Eds.; Today & Tomorrow’s Printers and Publ.: Delhi, India, 2004; pp. 141–187. [Google Scholar]
- IRRI. Standard Evaluation System for Rice, 5th ed.; International Rice Research Institute: Los Baños, CA, USA, 2013. [Google Scholar]
- Kumar, I.; Singh, T. A rapid method for identifying different dwarfing genes in rice. Rice Genet. Newsl. 1984, 1, 134–135. [Google Scholar]
- Murray, H.G.; Thompson, W.F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Ellis, M.; Spielmeyer, W. “Perfect” markers for the rice sd-1 semi-dwarfing gene. Int. Rice Res. Notes. 2002, 27, 13–14. [Google Scholar]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; obos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9829–9832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. Mapmaker: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Ellur, R.K.; Khanna, A.; Yadav, A.; Pathania, S.; Rajashekara, H.; Singh, V.K.; Gopalakrishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; et al. Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 2016, 242, 330–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Genotype | Control | GA3 Treated | Relative Response in Plant Height (%) | ||
|---|---|---|---|---|---|
| PH10 | PH18 | PH10 | PH18 | ||
| Pusa 1652 | 7.5 c | 18.7 c | 7.3 c | 27.1 c | 45.3 a |
| Chakhao Poireiton | 14.0 a | 30.8 a | 14.8 a | 37.7 a | 22.4 b |
| IR64 (sd1-d check) | 13.1 b | 21.8 b | 13.9 b | 30.9 b | 41.4 a |
| CD (p < 0.05) | 0.64 | 0.40 | 0.33 | 2.03 | 12.9 |
| Genotype | PHt | NPT | PnL | RPG (%) |
|---|---|---|---|---|
| BL 1 | 87.0 b | 16.0 a | 24.0 ab | 87.5 |
| BL 2 | 91.0 b | 17.0 a | 20.0 b | 87.5 |
| BL 3 | 103.0 b | 14.0 a | 25.0 ab | 91.6 |
| BL 4 | 97.0 b | 18.0 a | 21.0 b | 91.6 |
| Chakhao Poireiton | 153.2 a | 6.4 b | 28.0 a | - |
| Pusa 1652 | 87.6 b | 14.0 a | 19.8 b | - |
| CD (p < 0.05) | 16.81 | 4.22 | 5.44 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhuvaneswari, S.; Gopala Krishnan, S.; Ellur, R.K.; Vinod, K.K.; Bollinedi, H.; Bhowmick, P.K.; Bansal, V.P.; Nagarajan, M.; Singh, A.K. Discovery of a Novel Induced Polymorphism in SD1 Gene Governing Semi-Dwarfism in Rice and Development of a Functional Marker for Marker-Assisted Selection. Plants 2020, 9, 1198. https://doi.org/10.3390/plants9091198
Bhuvaneswari S, Gopala Krishnan S, Ellur RK, Vinod KK, Bollinedi H, Bhowmick PK, Bansal VP, Nagarajan M, Singh AK. Discovery of a Novel Induced Polymorphism in SD1 Gene Governing Semi-Dwarfism in Rice and Development of a Functional Marker for Marker-Assisted Selection. Plants. 2020; 9(9):1198. https://doi.org/10.3390/plants9091198
Chicago/Turabian StyleBhuvaneswari, Shivashankar, Subbaiyan Gopala Krishnan, Ranjith Kumar Ellur, Kunnummal Kurungara Vinod, Haritha Bollinedi, Prolay Kumar Bhowmick, Vijay Prakash Bansal, Mariappan Nagarajan, and Ashok Kumar Singh. 2020. "Discovery of a Novel Induced Polymorphism in SD1 Gene Governing Semi-Dwarfism in Rice and Development of a Functional Marker for Marker-Assisted Selection" Plants 9, no. 9: 1198. https://doi.org/10.3390/plants9091198