The Use of Nitrogen and Its Regulation in Cereals: Structural Genes, Transcription Factors, and the Role of miRNAs
Abstract
:1. Introduction
2. The Use of Nitrogen in Cereals
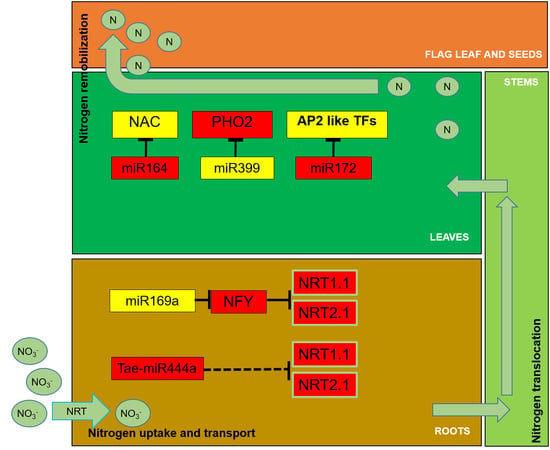
2.1. Nitrogen Uptake and Transport
2.2. Nitrogen Reduction and Assimilation
2.3. Nitrogen Translocation and Remobilization
3. Transcription Factors Regulating N Use in Cereals
4. MiRNAs and Target Genes Involved in the Regulation of N Use in Cereals
4.1. MiRNA Regulation of N Uptake
4.2. MiRNA Regulation of N Remobilization
4.3. Other N Stress Responsive miRNAs
| miRNA/Gene | Species Transformed | Genetic Modification | Genes Functionally Validated as miRNA Targets | Effects of the Transgenic/Knockout Gene or miRNA | Reference |
|---|---|---|---|---|---|
| Osa-miR393 | O. sativa | Overexpression and Knockout mutation | OsAFB2 and OsTB1 | Overexpression mimicked N-mediated tillering and knockout mutation repressed N-promoted tillering | [81] |
| OsDof18 | Knockout mutation | Reduction of the expression of ammonium transporter genes and ammonium uptake | [61] | ||
| Osa-miR528 | A. stolonifera | Transgenic expression | AAO, COPPER ION BINDING PROTEIN1 | Increasing of biomass, total N accumulation and chlorophyll synthesis, nitrite reductase activity and reduced AAO activity | [80] |
| Zma-miR528 | Knockdown mutation | ZmLACCASE3 (ZmLAC3) and ZmLACCASE5 (ZmLAC5) | Significant increasing of lignin content and rind penetrometer resistance of maize stems | [41] | |
| ZmLAC3 | Z. mays | Overexpression | Significant increasing of lignin content and rind penetrometer resistance of maize stems | [41] | |
| OsGS1 | O. sativa | Overexpression | Improving of N use efficiency | [80] | |
| HvGS1-1 | H. vulgare | Overexpression | Higher grain yields and NUE when grown under three different N supplies and two levels of atmospheric CO2. Improving of grain yield and NUE | [44] | |
| Tae-MIR444a | N. tabacum | Transgenic expression | NtNRT1.1-s, NtNET1.1-t, NtNRT2.1 and AEEs; NtCAT1;1, NtPOD1;3, and NtPOD4 | Increasing of N acquisition and cellular ROS detoxification in N-deprived plants | [76] |
| ZmDof1 | T. aestivum | Transgenic expression | Increasing biomass and yield. Down-regulation of genes involved in photosynthesis | [60] | |
| Zma-miR528 | Z. mays | Overexpression | ZmLAC3 and ZmLAC5 | Reduction of lignin biosynthesis under Nitrogen-Luxury Conditions | [41] |
| ZmGln1-3/ZmGln1-4 | Knockout mutation | Reduction of kernel size and kernel number | [42] | ||
| SbGln1 | S. bicolor | Overexpression | Greater tillering and up to 2.1-fold increase in shoot vegetative biomass under optimal nitrogen conditions | [43] | |
| ZmDof1 | O. sativa | Transgenic expression | Increasing of nitrogen assimilation and enhancing plant growth under low-nitrogen conditions | [59] | |
| ZmDof1 | S. bicolor | Transgenic expression | Increasing biomass and yield. Down-regulation of genes involved in photosynthesis | [60] | |
| Tae-miR2275 | N. tabacum | Transgenic expression and knockdown | TaPRP, TaBDP, TaWRK, TaSPK, TaPP, TaAAT, TaNTA, TaIM | Increasing of the biomass and N accumulation in overexpressing lines. Decreased biomass and plant N amount after N starvation in knockdown mutants | [83] |
| OsNRT2.3b | O. sativa | Overexpression | Increasing of N, Fe, and P uptake. Improving of the grain yield and nitrogen use efficiency (NUE) by 40% | [25] | |
| OsBT | O. sativa | Mutation | Increasing of NUE by 20% under low nitrogen conditions | [57] | |
| TaNFYA-B1 | T. aestivum | Overexpression | Significant increasing of both nitrogen and phosphorus uptake and grain yield under differing nitrogen and phosphorus supply levels | [73] |
5. MiRNAs and Crosstalk Between Nutrients in Cereals
6. Conclusions
| miRNA Families | Rice | Maize | Bread Wheat | Durum Wheat | Validated/Putative Target Genes | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L | R | L | R | L | R | L | R | ||||
| miR156 | Squamosa promoter binding protein-like (SBP-box) | [72,79,82] | |||||||||
| miR157 | * | [66] | |||||||||
| miR159 | MYB33, MYB65 | [63,72] | |||||||||
| miR160 | Auxin response, ARF22 | [65,68,70] | |||||||||
| miR162 | DCL1 | [72] | |||||||||
| miR164 | ^ | NAC, NAC7 | [65,68,70,72,79] | ||||||||
| miR166 | START domain containing protein, HD-Zip TFs | [68,75] | |||||||||
| miR167 | ^ | ^ | ARF8 | [65,67,68,72,79] | |||||||
| miR168 | ARGONAUTE1 | [65,79] | |||||||||
| miR169 | * | CCAAT-TF WHAP6, HAP2 like protein | [63,65,66,67,72,73,75] | ||||||||
| miR171 | Scarecrow-like TF; Protein FAN | [72] | |||||||||
| miR172 | AP2 like TFs, APETALA2, Bzip TF family protein | [63,65,72] | |||||||||
| miR319 | ^ | MYB and TCP transcriptional factors | [65,67] | ||||||||
| miR393 | AFB2 | [67,72] | |||||||||
| miR394 | F-box domain containing protein | [72] | |||||||||
| miR395 | APS1, APS4 | [65,72] | |||||||||
| miR396 | GRF TFs, rhodenase-like proteins, kinesin-like protein B | [72] | |||||||||
| miR397 | * | Laccase | [65,66,72] | ||||||||
| miR398 | * | COX | [65,66,72] | ||||||||
| miR399 | PHO2 | [65,67,68,70,72,82,95] | |||||||||
| miR408 | * | PLANTACYANIN | [65,66,72,75] | ||||||||
| miR415 | * | Aminoacylase; N-acyl-L-amino-acid amidohydrolase | [66] | ||||||||
| miR444 | ^ | MIKC-type MADS-box TFs, Maturase K, GRAS TFs | [64,67,68,76,82] | ||||||||
| miR528 | IAR1, CBP/OsDCL1, POD, SOD | [41,65,72,75,79] | |||||||||
| miR529 | Squamosa promoter binding protein-like (SBP-box) | [63] | |||||||||
| miR530 | Hairpin-induced protein 1 domain containing protein | [95] | |||||||||
| miR820 | DRM2 (DNA (cytosine-5)-methyltransferase) | [79] | |||||||||
| miR821 | GDH1 (Glutamate dehydrogenase) | [79] | |||||||||
| miR827 | * | ^ | SPX E3 ligase, CLP | [65,66,67,68] | |||||||
| miR1118 | [82] | ||||||||||
| miR1129 | [82] | ||||||||||
| miR1133 | Calmodulin-like, SET domain, early nodulin proteins, etc. | [82] | |||||||||
| miR1136 | [82] | ||||||||||
| miR1214 | * | [66] | |||||||||
| miR1318 | Calcium binding proteins or Calcium ATPases | [79] | |||||||||
| miR2199 | * | [66] | |||||||||
| miR2275 | * | PRP, BDP, WRK, SPK, PP, AAT, NTA, IM | [83] | ||||||||
| miR3979 | [63] | ||||||||||
| Down-regulated | |||||||||||
| Up-regulated | |||||||||||
| Different miRNA family members display different expression pattern | |||||||||||
| Different developmental stages display different expression pattern | |||||||||||
| * | Seedlings | ||||||||||
| ^ | Different behaivor in different crop varieties | ||||||||||
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ladha, J.K.; Tirol-Padre, A.; Reddy, C.K.; Cassman, K.G.; Verma, S.; Powlson, D.S.; Van Kessel, C.; Richter, D.D.B.; Chakraborty, D.; Pathak, H. Global nitrogen budgets in cereals: A 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 2016, 6, 19355. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, N.; Cavaletto, M. Cereals Proteins. In Proteins in Food Processing. A volume in Woodhead Publishing Series in Food Science, Technology and Nutrition, 2nd ed.; Yada, R.Y., Ed.; Elsevier: Kidlington, UK, 2018; pp. 223–244. [Google Scholar]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Jazmín Reyes-Hernández, B.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen fixation in cereals. Front Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef] [PubMed]
- Mulvaney, R.L.; Khan, S.A.; Ellsworth, T.R. Synthetic Nitrogen Fertilizers Deplete Soil Nitrogen: A Global Dilemma for Sustainable Cereal Production. J. Environ. Qual. 2009, 38, 2295. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Holtham, L.R.; Okamoto, M.; Garnett, T.P. Nitrate uptake and its regulation in relation to improving nitrogen use efficiency in cereals. Semin. Cell Dev. Boil. 2018, 74, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.; Felgate, H.; Watmough, N.; Thomson, A.; Baggs, E.; Watmough, N. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 2009, 27, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs AND THEIR REGULATORY ROLES IN PLANTS. Annu. Rev. Plant Boil. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Able, A.J.; Able, J.A. SMARTER De-Stressed Cereal Breeding. Trends Plant Sci. 2016, 21, 909–925. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Carbonell, A.; Marques, M.C.; Bustamante, A.; Fares, M.A.; Rodrigo, G.; Gomez, G. Inferring the regulatory network of the miRNA-mediated response to biotic and abiotic stress in melon. BMC Plant Boil. 2019, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Viswanathan, C.; Jianhua, Z.; Zhu, J.K. Smal RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Timmermans, M.C. Small RNAs are on the move. Nature 2010, 467, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and Interpretation of Factors Which Contribute to Efficiency of Nitrogen Utilization1. Agron. J. 1982, 74, 562. [Google Scholar] [CrossRef]
- Hansen, N.J.S.; Plett, D.; Berger, B.; Garnett, T. Tackling Nitrogen Use Efficiency in Cereal Crops Using High-Throughput Phenotyping. In Engineering Nitrogen Utilization in Crop Plants; Shrawat, A., Zayed, A., Lightfoot, D., Eds.; Springer Nature: Cham, Switzerland, 2018; pp. 121–139. [Google Scholar]
- Muchow, R. Nitrogen utilization efficiency in maize and grain sorghum. Field Crop. Res. 1998, 56, 209–216. [Google Scholar] [CrossRef]
- Presterl, T.; Seitz, G.; Landbeck, M.; Thiemt, W.; Schmidt, W.; Geiger, H.H. Improving nitrogen use efficiency in European maize: Estimation of quantitative parameters. Crop Sci. 2003, 43, 1259–1265. [Google Scholar] [CrossRef]
- Anbessa, Y.; Juskiw, P.; Good, A.; Nyachiro, J.; Helm, J. Genetic Variability in Nitrogen Use Efficiency of Spring Barley. Crop. Sci. 2009, 49, 1259. [Google Scholar] [CrossRef]
- Namai, S.; Toriyama, K.; Fukuta, Y. Genetic variations in dry matter production and physiological nitrogen use efficiency in rice (Oryza sativa L.) varieties. Breed. Sci. 2009, 59, 269–276. [Google Scholar] [CrossRef]
- Nehe, A.; Misra, S.; Murchie, E.; Chinnathambi, K.; Foulkes, M. Genetic variation in N-use efficiency and associated traits in Indian wheat cultivars. Field Crop. Res. 2018, 225, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.; Toubia, J.; Garnett, T.; Tester, M.; Kaiser, B.N.; Baumann, U. Dichotomy in the NRT Gene Families of Dicots and Grass Species. PLoS ONE 2010, 5, e15289. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hüner, N.; Tian, L. Identification and molecular characterization of the Brachypodium distachyon NRT2 family, with a major role of BdNRT2.1. Physiol. Plant 2019, 165, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Feng, H.; Yan, M.; Fan, X.; Shen, Q.; Miller, A.J.; Xu, G. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 2011, 62, 2319–2332. [Google Scholar] [Green Version]
- Cai, C.; Wang, J.Y.; Zhu, Y.G.; Shen, Q.R.; Li, B.; Tong, Y.P.; Li, Z.S. Gene Structure and Expression of the High-affinity Nitrate Transport System in Rice Roots. J. Integr. Plant Boil. 2008, 50, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2017, 113, 7118–7123. [Google Scholar] [CrossRef] [PubMed]
- Buchner, P.; Hawkesford, M.J. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J. Exp. Bot. 2014, 65, 5697–5710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabuchi, M.; Abiko, T.; Yamaya, T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Giehl, R.F.H.; Geldner, N.; Salt, D.E.; Von Wirén, N. Root zone–specific localization of AMTs determines ammonium transport pathways and nitrogen allocation to shoots. PLoS Biol. 2018, 16, e2006024. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Loqué, D.; Kojima, S.; Rauch, S.; Ishiyama, K.; Inoue, E.; Takahashi, H.; von Wirén, N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 2007, 19, 2636–2652. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.H.; Laginha, A.M.; Duan, F.; Rentsch, D.; Yuan, L.; von Wire’n, N. A critical role of AMT2;1 in root-toshoot translocation of ammonium in Arabidopsis. Mol. Plant 2017, 10, 1449–1460. [Google Scholar] [CrossRef]
- Loqué, D.; von Wirén, N. Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 2004, 55, 1293–1305. [Google Scholar] [CrossRef] [Green Version]
- Saiki, S.; Von Wirén, N.; Sonoda, Y.; Ikeda, A.; Yamaya, T.; Yamaguchi, J. Distinct Expression and Function of Three Ammonium Transporter Genes (OsAMT1;1—1;3) in Rice. Plant Cell Physiol. 2003, 44, 726–734. [Google Scholar]
- Kumar, A.; Silim, S.N.; Okamoto, M.; Siddiqi, M.Y.; Glass, A.D.M. Differential expression of three members of the AMT1 gene family encoding putative high affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ. 2003, 26, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, A.; Moriya, K.; Sonoda, Y.; Ikeda, A.; von Wirén, N.; Hayakawa, T.; Yamaguchi, J.; Yamaya, T. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 2003, 44, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Huss-Danell, K.; Högberg, P.; Näsholm, T.; Huss-Danell, K. Uptake of glycine by field grown wheat. New Phytol. 2001, 150, 59–63. [Google Scholar]
- Biernath, C.; Fischer, H.; Kuzyakov, Y. Root uptake of N-containing and N-free low molecular weight organic substances by maize: A 14C/15N tracer study. Soil Boil. Biochem. 2008, 40, 2237–2245. [Google Scholar] [CrossRef]
- Meyer, C.; Stitt, M. Nitrate Reduction and signalling. In Plant Nitrogen; Lea, P.J., Morot-Gaudry, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 37–59. [Google Scholar]
- Andrews, M.; Lea, P.J.; Raven, J.A.; Lindsey, K. Can genetic manipulation of plant nitrogen assimilation enzymes result in increased crop yield and greater N-use efficiency? An assessment. Ann. Appl. Boil. 2004, 145, 25–40. [Google Scholar] [CrossRef]
- Brauer, E.K.; Rochon, A.; Bi, Y.M.; Bozzo, G.G.; Rothstein, S.J.; Shelp, B.J. Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant. 2011, 141, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Habash, D.Z.; Massiah, A.J.; Rong, H.L.; Wallsgrove, R.M.; Leigh, R.A. The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Boil. 2001, 138, 83–89. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W.X. MicroRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatou, C.; Balliau, T.; Valot, B.; Davanture, M.; Dubois, F.; et al. Two cytosolic glutamine synthetase isoforms of maize (Zea mays L.) are specifically involved in the control of grain production. Plant Cell 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Urriola, J.; Rathore, K.S. Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res. 2015, 24, 397. [Google Scholar] [CrossRef]
- Gao, Y.; de Bang, T.C.; Schjoerring, J.K. Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 2019, 17, 1209–1221. [Google Scholar] [PubMed]
- Curci, P.L.; Bergès, H.; Marande, W.; Maccaferri, M.; Tuberosa, R.; Sonnante, G. Asparagine synthetase genes (AsnS1 and AsnS2) in durum wheat: Structural analysis and expression under nitrogen stress. Euphytica 2018, 214, 36. [Google Scholar] [CrossRef]
- Hayakawa, T.; Nakamura, T.; Hattori, F.; Mae, T.; Ojima, K.; Yamaya, T. Cellular localization of NADH-dependent glutamate-synthase protein in vascular bundles of unexpanded leaf blades and young grains of rice plants. Planta 1994, 193, 455–460. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Porto, M.; Pinheiro, M.; Batista, V.; dos Santos, R.; de Albuquerque Melo Filho, P.; de Lima, L.M. Plant promoters: An approach of structure and function. Mol. Biotechnol. 2014, 56, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [Green Version]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef]
- Guan, P.; Ripoll, J.J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Wang, R.; Zhao, L.; Zhang, C.; Li, Z.; Lei, Z.; Liu, F.; Guan, P.; Chu, Z.; Crawford, N.M.; et al. The Arabidopsis NRG2 Protein Mediates Nitrate Signaling and Interacts with and Regulates Key Nitrate Regulators. Plant Cell 2016, 28, 485–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araus, V.; Vidal, E.A.; Puelma, T.; Álamos, S.; Mieulet, D.; Guiderdoni, E.; Gutiérrez, R.A. Members of BTB Gene Family of Scaffold Proteins Suppress Nitrate Uptake and Nitrogen Use Efficiency1. Plant Physiol. 2016, 171, 1523–1532. [Google Scholar] [PubMed]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef]
- Kurai, T.; Wakayama, M.; Abiko, T.; Yanagisawa, S.; Aoki, N.; Ohsugi, R. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 2011, 9, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Peña, P.A.; Quach, T.; Sato, S.; Ge, Z.; Nersesian, N.; Changa, T.; Dweikat, I.; Soundararajan, M.; Clemente, T.E. Expression of the Maize Dof1 Transcription Factor in Wheat and Sorghum. Front. Plant Sci. 2017, 8, 434. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Yang, W.; Wei, J.; Yoon, H.; An, G. Transcription Factor OsDOF18 Controls Ammonium Uptake by Inducing Ammonium Transporters in Rice Roots. Mol. Cells 2017, 40, 178–185. [Google Scholar] [Green Version]
- Curci, P.L.; Cigliano, R.A.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 7, 1141. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Wang, W.; Zhang, Z.; Liang, Y.; Gao, X.; Li, P.; Liu, Y.; Zhang, L.; Chu, C. Identification of microRNAs in rice root in response to nitrate and ammonium. J. Genet. Genom. 2016, 43, 651–661. [Google Scholar] [CrossRef]
- Shin, S.Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genom. 2018, 19, 532. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome-Wide Identification of MicroRNAs in Response to Low Nitrate Availability in Maize Leaves and Roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Z.; Yang, C.; Yang, Z.; Li, H.; Wu, Y. Physiological responses and small RNAs changes in maize under nitrogen deficiency and resupply. Genes Genom. 2019, 41. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; De Paola, D.; Janni, M.; Curci, P.L.; Sonnante, G. Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS ONE 2017, 12, e0183253. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; Liuzzi, V.; Curci, P.L.; Sonnante, G. MicroRNAs in durum wheat seedlings under chronic and short-term nitrogen stress. Funct. Integr. Genom. 2018, 18, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Colaiacovo, M.; Subacchi, A.; Bagnaresi, P.; Lamontanara, A.; Cattivelli, L.; Faccioli, P. A computational-based update on microRNAs and their targets in barley (Hordeum vulgare L.). BMC Genom. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Rani, M.; Bansal, N.; Gayatri; Venkatesh, K.; Mandal, P.K. Nitrate Starvation Induced Changes in Root System Architecture, Carbon:Nitrogen Metabolism, and miRNA Expression in Nitrogen-Responsive Wheat Genotypes. Appl. Biochem. Biotechnol. 2015, 177, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ding, H.; Zhu, J.K.; Zhang, F.; Li, W.X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef]
- Zhao, M.; Tai, H.; Sun, S.; Zhang, F.; Xu, Y.; Li, W.X. Cloning and characterization of maize miRNA involved in response to nitrogen deficiency. PLoS ONE 2012, 7, e29669. [Google Scholar] [CrossRef]
- Qu, B.; He, X.; Wang, J.; Zhao, Y.; Teng, W.; Shao, A.; Zhao, X.; Ma, W.; Wang, J.; Li, B.; et al. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 2015, 167, 411–423. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, Z.; Mo, Q.; Zou, C.; Li, W.; Xu, Y.; Xie, C. Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Ann. Bot. 2013, 112, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, S.; Nonis, A.; Begheldo, M.; Manoli, A.; Palme, K.; Caporale, G.; Ruperti, B.; Quaggiotti, S. Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 2012, 35, 1137–1155. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, C.; Zhang, Y.; Zhang, F.; Du, X.; Gu, J.; Xiao, K. Wheat microRNA Member TaMIR444a Is Nitrogen Deprivation-Responsive and Involves Plant Adaptation to the Nitrogen-Starvation Stress. Plant Mol. Boil. Rep. 2016, 34, 931–946. [Google Scholar] [CrossRef]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Uauy, C.; Dubcovsky, J.; Grusak, M.A. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 2009, 60, 4263–4274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nischal, L.; Mohsin, M.; Khan, I.; Kardam, H.; Wadhwa, A.; Abrol, Y.P.; Iqbal, M.; Ahmad, A. Identification and Comparative Analysis of MicroRNAs Associated with Low-N Tolerance in Rice Genotypes. PLoS ONE 2012, 7, e50261. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive Expression of Rice MicroRNA528 Alters Plant Development and Enhances Tolerance to Salinity Stress and Nitrogen Starvation in Creeping Bentgrass1. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xia, K.; Liang, Z.; Chen, K.; Gao, C.; Zhang, M. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 2016, 6, 32158. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Guo, L.; Lu, W.; Li, X.; Chen, H.; Guo, C.; Xiao, K. Expression pattern analysis of microRNAs in root tissue of wheat (Triticum aestivum L.) under normal nitrogen and low nitrogen conditions. J. Plant Biochem. Biotechnol. 2015, 24, 143–153. [Google Scholar] [CrossRef]
- Qiao, Q.; Wang, X.; Yang, M.; Zhao, Y.; Gu, J.; Xiao, K. Wheat miRNA member TaMIR2275 involves plant nitrogen starvation adaptation via enhancement of the N acquisition-associated process. Acta Physiol. Plant. 2018, 40, 183. [Google Scholar] [CrossRef]
- Liang, G.; Ai, Q.; Yu, D. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci. Rep. 2015, 5, 11813. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A.; Blatt, M.R. Regulation of macronutrient transport. New Phytol. 2009, 181, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.H. Nitrogen Limitation Adaptation Recruits 847 Phosphate2 to Target the Phosphate Transporter PT2 for Degradation during the Regulation of 848 Arabidopsis Phosphate Homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Boil. 2007, 65, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Hannam, C.; Gu, H.; Bi, Y.M.; Rothstein, S.J. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007, 50, 320–337. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in Arabidopsis. PLoS Genet. 2011, 7, 1002021. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA Involved in Phosphate-Starvation Response in Arabidopsis. Curr. Boil. 2005, 15, 2038–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. pho2, a Phosphate Overaccumulator, Is Caused by a Nonsense Mutation in a MicroRNA399 Target Gene1. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, MicroRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants1. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.l. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Lu, Y.; Xie, W.; Zhu, T.; Lian, X. Transcriptome response to nitrogen starvation in rice. J. Biosci. 2012, 37, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ball, G.; Hodgman, C.; Coules, A.; Zhao, H.; Lu, C. Analysis of Gene Regulatory Networks of Maize in Response to Nitrogen. Genes 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuluaga, D.L.; Sonnante, G. The Use of Nitrogen and Its Regulation in Cereals: Structural Genes, Transcription Factors, and the Role of miRNAs. Plants 2019, 8, 294. https://doi.org/10.3390/plants8080294
Zuluaga DL, Sonnante G. The Use of Nitrogen and Its Regulation in Cereals: Structural Genes, Transcription Factors, and the Role of miRNAs. Plants. 2019; 8(8):294. https://doi.org/10.3390/plants8080294
Chicago/Turabian StyleZuluaga, Diana L., and Gabriella Sonnante. 2019. "The Use of Nitrogen and Its Regulation in Cereals: Structural Genes, Transcription Factors, and the Role of miRNAs" Plants 8, no. 8: 294. https://doi.org/10.3390/plants8080294