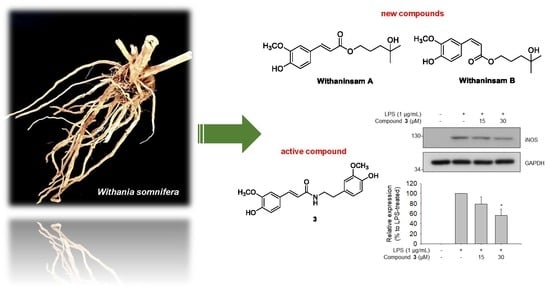
Withaninsams A and B: Phenylpropanoid Esters from the Roots of Indian Ginseng (Withania somnifera)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Compounds
2.2. Structure Elucidation of Compounds
2.3. Inhibitory Effects of Compounds 1–6 on LPS-Induced NO Production in RAW 264.7 Cells
3. Materials and Methods
3.1. Plant Material
3.2. Extraction and Isolation
Withaninsams A (1) and B (2)
3.3. Cell Viability Assay
3.4. NO Production Assay
3.5. TNF-α ELISA
3.6. Western Blotting
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Khare, C.P. Encyclopedia of Indian Medicinal Plants; Springer Verlag: New York, NY, USA, 2004; pp. 480–483. [Google Scholar]
- Gupta, G.L.; Rana, A.C. Withaniasomnifera (Ashwagandha): A review. Pharmacogn. Rev. 2007, 1, 129–136. [Google Scholar]
- Kim, S.; Yu, J.S.; Lee, J.Y.; Choi, S.U.; Lee, J.; Kim, K.H. Cytotoxic Withanoildes from the Roots of Indian Ginseng (Withaniasomnifera). J. Nat. Prod. 2019, 82, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.K.; Dhir, A. Withaniasomnifera: An Indian ginseng. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2008, 32, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Saxena, A.K.; Chandan, B.K.; Gupta, D.K.; Bhutani, K.K.; Anand, K.K. Adaptogenic activity of novel, withanolide-free aqueous fraction from the roots of Withaniasomnifera Dun. Phytother. Res. 2001, 15, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.A.; Weiner, J. Herbs That Heal: Prescription for Herbal Healing; Atrium Publishers Group: Mill Valley, CA, USA, 1994; p. 70. [Google Scholar]
- Vedi, M.; Rasool, M.; Sabina, E.P. Amelioration of bromobenzene hepatotoxicity by Withaniasomnifera pretreatment: Role of mitochondrial oxidative stress. Toxicol. Rep. 2014, 1, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Ziegenfuss, T.N.; Kedia, A.W.; Sandrock, J.E.; Raub, B.J.; Kerksick, C.M.; Lopez, H.L. Effects of an Aqueous Extract if Withaniasomnifera on Strength Training Adaptations and Recovery: The STAR Trial. Nurtrients 2018, 10, 1807. [Google Scholar] [CrossRef]
- Raghavan, A.; Shah, Z.A. Withaniasomnifera Improves Ischemic Stroke Outcomes by Attenuating PARP1-AIF-Mediated Caspase-Independent Apoptosis. Mol. Neurobiol. 2015, 52, 1093–1105. [Google Scholar] [CrossRef]
- Ma, T.; Zhang, W.-N.; Yang, L.; Zhang, C.; Lin, R.; Shan, S.-M.; Zhu, M.-D.; Luo, J.-G.; Kong, L.-Y. Cytotoxic withanolides from Physalis angulata var. villosa and the apoptosis-inducing effect via ROS generation and the activation of MAPK in human osteosarcoma cells. RSC Adv. 2016, 6, 53089–53100. [Google Scholar] [CrossRef]
- Habtemariam, S. Cytotoxicity and Immunosuppressive Activity of Withanoliedes from Discopodiumpenninervium. Planta Med. 1997, 63, 15–17. [Google Scholar] [CrossRef]
- Budhiraja, R.D.; Sudhir, S.; Garg, K.N. Antiinflmmatory Activity of 3 β-Hydroxy-2,3-dihydro-withanolide F. Planta Med. 1984, 50, 134–136. [Google Scholar] [CrossRef]
- Chakraborti, S.K.; De Barun, K.; Bandyopadhyay, T. Variations in the antitumor constituents of Withaniasomnifera dunal. Experientia 1974, 30, 852–853. [Google Scholar] [CrossRef] [PubMed]
- Mareggiani, G.; Picollo, M.I.; Zerba, E.; Burton, G.; Tettamanzi, M.C.; Benedetti-Doctorovich, M.O.V.; Veleiro, A.S. Antifeedant Activity of Withanolides from Salpichroaoriganifolia on Musca Domestica. J. Nat. Prod. 2000, 63, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.V.; Leiva Gonzalez, S.; Barboza, G.E.; Careaga, V.P.; Calvo, J.C.; Sacca, P.A.; Nicotra, V.E. Phytochemical Study of the Genus Salpichroa (Solanaceae), Chemotaxonomic Considerations, and Biological Evaluation in Prostate and Breast Cancer Cells. Chem. Biodivers. 2017, 14, e1700118. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, C.-M.; Gallagher, R.J.; Timmermann, B.N. Antiproliferative withanolides from several solanaceous spicies. Nat. Prod. Res. 2014, 28, 1941–1951. [Google Scholar] [CrossRef]
- Bolleddula, J.; Fitch, W.; Vareed, S.K.; Nair, M.G. Identification of metabolies in Withaniasomnifera fruits by liquid chromatography and high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 1277–1290. [Google Scholar] [CrossRef]
- Elsakka, M.; Grigorescu, E.; Stănescu, U.; Dorneanu, V. New data referring to chemistry of Withaniasomnifera species. Rev. Med. Chir. Soc. Med. Nat. IASI 1990, 94, 385–387. [Google Scholar]
- Jayaprakasam, B.; Zhang, Y.; Seeram, N.P.; Nair, M.G. Growth inhibition of human tumor cell lines by withanolides from Withaniasomnifera leaves. Life Sci. 2003, 74, 125–132. [Google Scholar] [CrossRef]
- So, H.M.; Eom, H.J.; Lee, D.; Kim, S.; Kang, K.S.; Lee, I.K.; Baek, K.-H.; Park, J.Y.; Kim, K.H. Bioactivity evaluations of betulin identified from the bark of Betula platyphylla var. japonica for cancer therapy. Arch. Pharm. Res. 2018, 41, 815–822. [Google Scholar] [CrossRef]
- Yu, J.S.; Roh, H.-S.; Baek, K.-H.; Lee, S.; Kim, S.; So, H.M.; Moon, E.; Pang, C.; Jang, T.S.; Kim, K.H. Bioactivity-guided isolation of ginsenosides from Korean Red Ginsengwith cytotoxic activity against human lung adenocarcinoma cells. J. Ginseng Res. 2018, 42, 562–570. [Google Scholar] [CrossRef]
- Baek, S.C.; Choi, E.; Eom, H.J.; Jo, M.S.; Kim, S.; So, H.M.; Kim, S.H.; Kang, K.S.; Kim, K.H. LC/MS-based analysis of bioactive compounds from the bark of Betula platyphylla var. japonica and their effects on regulation of adipocyte and osteoblast differentiation. Nat. Prod. Sci. 2018, 24, 235–240. [Google Scholar] [CrossRef]
- Yu, J.S.; Lee, D.; Lee, S.R.; Lee, J.W.; Choi, C.-I.; Jang, T.S.; Kang, K.S.; Kim, K.H. Chemical characterization of cytotoxic indole acetic acid derivative from Mulberry fruit (Morus alba L.) against human cervical cancer. Bioorg. Chem. 2018, 76, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Pavia, D.L.; Lampman, G.M.; Kriz, G.S.; Vyvyan, J.R. Introduction to Spectroscopy, 4th ed.; Cengage Learning: Stamford, CT, USA, 2008; pp. 237–297. [Google Scholar]
- Faig, J.J.; Klein, S.; Ouimet, M.A.; Yu, W.; Uhrich, K.E. Attenuating Oxidative Stress Via Oxalate Ester-Containing Ferulic Acid-Based Poly(anhydride-esters) that Scavenge Hydrogen Peroxide. Macromol. Chem. Phys. 2016, 217, 108–114. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Y.; Zhou, Y.; Li, L.; Zhang, T.Y.; Zhang, H. Imidazolylidene carbene ligated palladium catalysis of the Heck reaction in the presence of air. Org. Biomol. Chem. 2003, 1, 3227–3231. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Min, H.; Jeong, Y.H.; Lee, S.K.; Lee, N.S.; Seo, E. Cytotoxic Constituents from the Whole Plant of Corydalis pallida. Arch. Pharm. Res. 2005, 28, 122–1227. [Google Scholar]
- Ma, J.; Jones, S.H.; Hecht, S.M. Phenolic acid amides: A new type of DNA strand scission agent from Piper caninum. Bioorg. Med. Chem. 2004, 12, 3885–3889. [Google Scholar] [CrossRef]
- Majumder, S.; Gipson, K.R.; Staples, R.J.; Odom, A.L. Pyrazole Synthesis Using a Titanium-Catalyzed multicomponent Coupling Reaction and Synthesis of Withasomnine. Adv. Synth. Catal. 2009, 351, 2013–2023. [Google Scholar] [CrossRef]
- Bao, K.; Fan, A.; Zhang, L.; Zhang, W.; Cheng, M.; Yao, X. Selective demethylation and debenzylation of aryl ethers by magnesium iodide under solvent-free conditions and its application to the total synthesis of natural products. Org. Biomol. Chem. 2009, 7, 5084–5090. [Google Scholar] [CrossRef]

 ) and key heteronuclear multiple bond correlation (HMBC) (
) and key heteronuclear multiple bond correlation (HMBC) (  ) correlations for compounds 1 and 2.
) correlations for compounds 1 and 2.
 ) and key heteronuclear multiple bond correlation (HMBC) (
) and key heteronuclear multiple bond correlation (HMBC) (  ) correlations for compounds 1 and 2.
) correlations for compounds 1 and 2.


| Position | 1 | 2 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 1 | 127.0 | 127.3 | ||
| 2 | 7.01 d (1.5) | 109.2 | 7.74 d (2.0) | 112.6 |
| 3 | 146.5 | 145.9 | ||
| 4 | 147.9 | 147.0 | ||
| 5 | 6.89 d (8.0) | 114.6 | 6.86 d (8.0) | 113.7 |
| 6 | 7.05 dd (1.5, 8.0) | 123.1 | 7.08 dd (2.0, 8.0) | 125.5 |
| 7 | 7.58 d (16.0) | 114.6 | 6.77 d (13.0) | 143.7 |
| 8 | 6.27 d (16.0) | 115.6 | 5.79 d (13.0) | 116.8 |
| 9 | 167.3 | 166.7 | ||
| 1′ | 4.16 t (7.0) | 64.4 | 4.09 t (7.0) | 64.4 |
| 2′ | 1.37 m | 28.6 | 1.30 m | 28.5 |
| 3′ | 1.67 m | 25.9 | 1.62 m | 25.8 |
| 4′ | n.d.b | n.d. b | ||
| 5′ | 1.23 s | 29.5 | 1.23 s | 29.5 |
| 6′ | 1.23 s | 29.5 | 1.23 s | 29.5 |
| –OCH3 | 3.91 s | 55.6 | 3.91 s | 55.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, S.C.; Lee, S.; Kim, S.; Jo, M.S.; Yu, J.S.; Ko, Y.-J.; Cho, Y.-C.; Kim, K.H. Withaninsams A and B: Phenylpropanoid Esters from the Roots of Indian Ginseng (Withania somnifera). Plants 2019, 8, 527. https://doi.org/10.3390/plants8120527
Baek SC, Lee S, Kim S, Jo MS, Yu JS, Ko Y-J, Cho Y-C, Kim KH. Withaninsams A and B: Phenylpropanoid Esters from the Roots of Indian Ginseng (Withania somnifera). Plants. 2019; 8(12):527. https://doi.org/10.3390/plants8120527
Chicago/Turabian StyleBaek, Su Cheol, Seoyoung Lee, Sil Kim, Mun Seok Jo, Jae Sik Yu, Yoon-Joo Ko, Young-Chang Cho, and Ki Hyun Kim. 2019. "Withaninsams A and B: Phenylpropanoid Esters from the Roots of Indian Ginseng (Withania somnifera)" Plants 8, no. 12: 527. https://doi.org/10.3390/plants8120527