Genome-Wide Identification and Expression Analyses of the Fibrillin Family Genes Suggest Their Involvement in Photoprotection in Cucumber
Abstract
:1. Introduction
2. Results
2.1. FBN Family Genes in Cucumber Genomes
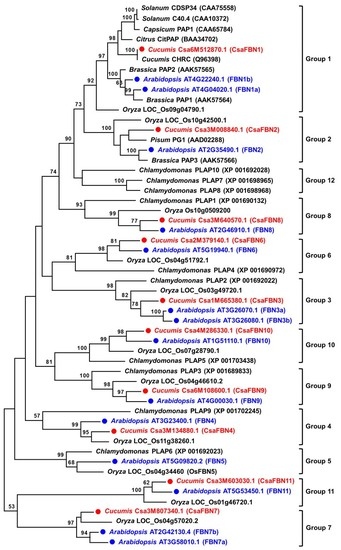
2.2. Phylogenetic Relationships among Cucumber FBN Genes
2.3. Gene Structure and Polymorphisms of FBN Genes in Cucumber
2.4. Expression Patterns of Cucumber FBNs in Various Tissues
2.5. Expression of Cucumber FBNs under High Light Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of the FBN Genes in Cucumber Genomes
4.3. Phylogenetic Analysis of FBN Genes
4.4. Sequence Polymorphism Analysis of the FBN Genes in Two Cucumber Varieties
4.5. Quantitative Real-Time RT-PCR Analysis
4.6. Measurement of Photosynthetic Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FBN | Fibrillin |
| PAPs | Plastid lipid-associated proteins |
| PGL | Plastoglobulin |
References
- Pozueta-Romero, J.; Rafia, F.; Houlne, G.; Cheniclet, C.; Carde, J.P.; Schantz, M.L.; Schantz, R. A Ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol. 1997, 115, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Kessler, F.; Schnell, D.; Blobel, G. Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 1999, 208, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Ytterberg, A.J.; Peltier, J.B.; van Wijk, K.J. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006, 140, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Gaffé, J.; Alcaraz, J.-P.; Carde, J.-P.; Bramley, P.M.; Fraser, P.D.; Kuntz, M. Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 2007, 68, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X.; Tice, A.B.; Pham, C.; Gantt, E. Inactivation of genes encoding plastoglobulin-like proteins in Synechocystis sp. PCC 6803 leads to a light-sensitive phenotype. J. Bacteriol. 2010, 192, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Lohscheider, J.N.; Rio Bartulos, C. Plastoglobules in algae: A comprehensive comparative study of the presence of major structural and functional components in complex plastids. Mar. Genom. 2016, 28, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Hadjeb, N.; Price, C.A. Synthesis of two chromoplast-specific proteins during fruit development in Capsicum annuum. Plant Physiol. 1989, 91, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Deruère, J.; Römer, S.; d’Harlingue, A.; Backhaus, R.A.; Kuntz, M.; Camara, B. Fibril assembly and carotenoid overaccumulation in chromoplasts: A model for supramolecular lipoprotein structures. Plant Cell 1994, 6, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Winkenbach, F.; Falk, H.; Liedvogel, B.; Sitte, P. Chromoplasts of Tropaeolum majus L.: Isolation and characterization of lipoprotein elements. Planta 1976, 128, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Knoth, R.; Hansmann, P.; Sitte, P. Chromoplasts of Palisota barteri, and the molecular structure of chromoplast tubules. Planta 1986, 168, 167–174. [Google Scholar] [PubMed]
- Vishnevetsky, M.; Ovadis, M.; Itzhaki, H.; Levy, M.; Libal-Weksler, Y.; Adam, Z.; Vainstein, A. Molecular cloning of a carotenoid-associated protein from Cucumis sativus corollas: Homologous genes involved in carotenoid sequestration in chromoplasts. Plant J. 1996, 10, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Pruvot, G.; Cuiné, S.; Peltier, G.; Rey, P. Characterization of a novel drought-induced 34-kDa protein located in the thylakoids of Solanum tuberosum L. plants. Planta 1996, 198, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Langenkämper, G.; Manac’h, N.; Broin, M.; Cuiné, S.; Becuwe, N.; Kuntz, M.; Rey, P. Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in Solanaceae and in response to drought in other species. J. Exp. Bot. 2001, 52, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Wu, S.S.H.; Ratnayake, C.; Huang, A.H.C. Brassica rapa has three genes that encode proteins associated with different neutral lipids in plastids of specific tissues. Plant Physiol. 2001, 126, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; McNellis, T.W. Fibrillin protein function: The tip of the iceberg? Trends Plant Sci. 2011, 16, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Laizet, Y.; Pontier, D.; Mache, R.; Kuntz, M. Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J. Genome Sci. Technol. 2004, 3, 19–28. [Google Scholar] [CrossRef]
- Lundquist, P.K.; Poliakov, A.; Bhuiyan, N.H.; Zybailov, B.; Sun, Q.; van Wijk, K.J. The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 2012, 158, 1172–1192. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Laremore, T.N.; Smith, P.B.; Maximova, S.N.; McNellis, T.W. Knockdown of FIBRILLIN4 gene expression in apple decreases plastoglobule plastoquinone content. PLoS ONE 2012, 7, e47547. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Maximova, S.N.; Jensen, P.J.; Lehman, B.L.; Ngugi, H.K.; McNellis, T.W. FIBRILLIN4 is required for plastoglobule development and stress resistance in apple and Arabidopsis. Plant Physiol. 2010, 154, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sulpice, R.; Himmelbach, A.; Meinhard, M.; Christmann, A.; Grill, E. Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 2006, 103, 6061–6066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youssef, A.; Laizet, Y.; Block, M.A.; Marechal, E.; Alcaraz, J.P.; Larson, T.R.; Pontier, D.; Gaffe, J.; Kuntz, M. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 2010, 61, 436–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidi, P.A.; Kanwischer, M.; Baginsky, S.; Austin, J.R.; Csucs, G.; Dormann, P.; Kessler, F.; Brehelin, C. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 2006, 281, 11225–11234. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Lee, D.W.; Lee, K.R.; Jung, S.J.; Jeon, J.S.; Kim, H.U. Conserved function of fibrillin5 in the plastoquinone-9 biosynthetic pathway in Arabidopsis and rice. Front. Plant Sci. 2017, 8, 1197. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Lee, Y.; Kim, H.U. Fibrillin 5 is essential for plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases in Arabidopsis. Plant Cell 2015, 27, 2956–2971. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lough, T.J.; Lucas, W.J. Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 2006, 57, 203–232. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS ONE 2012, 7, e47576. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Liu, W.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Comprehensive analysis of the homeodomain-leucine zipper IV transcription factor family in Cucumis sativus. Genome 2013, 56, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Gan, D.; Zhuang, D.; Ding, F.; Yu, Z.; Zhao, Y. Identification and expression analysis of primary auxin-responsive Aux/IAA gene family in cucumber (Cucumis sativus). J. Genet. 2013, 92, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.A.; Appiano, M.; Bijsterbosch, G.; Visser, R.G.F.; Schouten, H.J.; Bai, Y. Functional characterization of cucumber (Cucumis sativus L.) Clade V MLO genes. BMC Plant. Biol. 2017, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.P. Effect of light and chilling temperatures on chilling-sensitive and chilling-resistant plants. Pretreatment of cucumber and spinach thylakoids in vivo and in vitro. Plant Physiol. 1977, 59, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, R.A.J.; Orr, G.R.; Raison, J.K. Inhibition of photosynthesis by chilling in the light. Plant Sci. 1987, 49, 75–79. [Google Scholar] [CrossRef]
- Chen, M.; Thelen, J.J. ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 1430–1444. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Thelen, J.J. Acyl-lipid desaturase 1 primes cold acclimation response in Arabidopsis. Physiol. Plant 2016, 158, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Critchley, C. Studies on the Mechanism of Photoinhibition in Higher Plants: I. effects of high light intensity on chloroplast activities in cucumber adapted to low light. Plant Physiol. 1981, 67, 1161–1165. [Google Scholar] [CrossRef] [PubMed]
- Heinnickel, M.L.; Grossman, A.R. The GreenCut: Re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth. Res. 2013, 116, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Vishnevetsky, M.; Ovadis, M.; Itzhaki, H.; Vainstein, A. CHRC, encoding a chromoplast-specific carotenoid-associated protein, is an early gibberellic acid-responsive gene. J. Biol. Chem. 1997, 272, 24747–24750. [Google Scholar] [CrossRef] [PubMed]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta (BBA) Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Gene ID | Location on Chromosome (5) | CDS Length (bp) | Protein (aa) | Fibrillin Domain | Best Arabidopsis Homologue | % ID | E-Value |
|---|---|---|---|---|---|---|---|---|
| (IPR006843) (Cdd:pfam04755) | ||||||||
| CsaFBN1 | Csa6M512870.1 (1) (Cucsa.044900.1 (2)) | Chr6 (26485594..26487860) | 969 | 323 | 98–312 | AT4G04020.1 (FBN1a, FIB1a, AtPGL35) | 60 | 5.1 × 10−96 |
| CsaFBN2 | Csa3M008840.1 (Cucsa.322030.1) | Chr3 (992602..995095) | 1092 | 364 | 139–353 | AT2G35490.1 (FBN2, FIB2, AtPGL40) | 51 | 5.7 × 10−77 |
| CsaFBN3 | Csa1M665380.1 (Cucsa.027640.1) | Chr1 (26931385..26939153) | 732 | 244 | 74–233 | AT3G26070.1 (FBN3a, FIB3a) | 69 | 2.0 × 10−71 |
| CsaFBN4 | Csa3M134880.1 (Cucsa.255550.1) | Chr3 (9072862..9075038) | 870 | 290 | 91–286 | AT3G23400.1 (FBN4, FIB4) | 62 | 1.0 × 10−83 |
| CsaFBN6 | Csa2M379140.1 (Cucsa.161880.1) | Chr2 (19193933..19197291) | 747 | 249 | 79–244 | AT5G19940.1 (FBN6, FIB6) | 58 | 1.3 × 10−72 |
| CsaFBN7 | Csa3M807340.1 (Cucsa.242700.1) | Chr3 (30901949..30909200) | 912 | 304 | 97–280 | AT2G42130.4 (FBN7b, FIB7b) | 72 | 2.2 × 10−97 |
| Csa3M807340.2 (As (3)) | Chr3 (30901949..30907204)(As) | 819 (As) | 273 (As) | 97–243 | AT2G42130.1 (FBN7b, FIB7b) | 72 | 8.1 × 10−82 | |
| CsaFBN8 | Csa3M640570.1 (Cucsa.149720.1) | Chr3 (25041843..25045492) | 837 | 279 | 58–270 | AT2G46910.1 (FBN8, FIB8) | 63 | 1.6 × 10−76 |
| Csa3M640570.2 (As) | Chr3 (25042753..25045492)(As) | 636 (As) | 212 (As) | 58–194 | AT2G46910.1 (FBN8, FIB8) | 66 | 2.6 × 10−53 | |
| CsaFBN9 | Csa6M108600.1 (Cucsa.120630.1) | Chr6 (7303911..7307440) | 642 | 214 | 39–204 | AT4G00030.1 (FBN9, FIB9) | 77 | 2.2 × 10−72 |
| CsaFBN10 | Csa4M286330.1 (Cucsa.153490.1) | Chr4 (11046106..11050268) | 1311 | 437 | 99–275 | AT1G51110.1 (FBN10, FIB10) | 67 | 7.2 × 10−129 |
| CsaFBN11 | Csa3M603030.1 (Cucsa.040680.1 (4)) | Chr3 (23336459..23340819) | 1773 | 590 | 359–443 | AT5G53450.1 (FBN11, AtFib11) | 66 | 9.3 × 10−216 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.; Lee, S.-C.; Kim, E.-H.; Song, K.; Yang, T.-J.; Kim, H.U. Genome-Wide Identification and Expression Analyses of the Fibrillin Family Genes Suggest Their Involvement in Photoprotection in Cucumber. Plants 2018, 7, 50. https://doi.org/10.3390/plants7030050
Kim I, Lee S-C, Kim E-H, Song K, Yang T-J, Kim HU. Genome-Wide Identification and Expression Analyses of the Fibrillin Family Genes Suggest Their Involvement in Photoprotection in Cucumber. Plants. 2018; 7(3):50. https://doi.org/10.3390/plants7030050
Chicago/Turabian StyleKim, Inyoung, Sang-Choon Lee, Eun-Ha Kim, Kihwan Song, Tae-Jin Yang, and Hyun Uk Kim. 2018. "Genome-Wide Identification and Expression Analyses of the Fibrillin Family Genes Suggest Their Involvement in Photoprotection in Cucumber" Plants 7, no. 3: 50. https://doi.org/10.3390/plants7030050