1. Introduction
India is the world’s 4th largest grower and producer of oil-producing crops, accounting for ~19% of worldwide acreage and 2.7% of production. Oilseed crops, just after cereals, play an important role in the Indian agricultural economy. India is on track to become the world’s third-largest consumer market and an importer of edible oils, meeting over 60% of its domestic consumption through imports at the cost of USD 10 billion per year [
1,
2,
3]. Domestic demand for edible oils and fats has been proliferating at 6% per year, but domestic output has only increased by 2% per year. The country’s significant scarcity of edible oils has been attributed to several issues, including the country’s ever-growing population, sudden climate change, rising household income, low productivity of oilseed crops, and a complicated disease–pest syndrome. Poor production performance of oilseed crops is the most important reason for India’s demand–supply mismatch in vegetable oils. Rapeseed–mustard is the third most extensively produced oilseed crop in India, accounting for ~32% of the country’s total oil pool [
4].
Indian mustard [
Brassica juncea (L.) Czern & Coss.] is the most widely cultivated oilseed crop in India, out of six economically important species of the rapeseed–mustard group, due to its greater sustainability to grow under diverse agro-climatic conditions [
5,
6,
7]. Natural amphiploid (2n = 4x = 36, AABB) India mustard is developed by natural crossing and genome doubling between two diploid progenitors,
Brassica campestris (2n = 2x = 20, AA) and
Brassica nigra (2n = 2x = 16, BB) [
8]. To meet the escalating demand for vegetable oils for India’s ever-increasing population, the productivity of Indian mustard must be increased. Even though this crop has achieved significant progress in terms of yield enhancement [
4], current production is insufficient to meet the country’s demand. The requisite productivity goals can be met by producing high-yielding hybrids, which is possible in this crop due to abundant heterosis for seed yield and its components and a stable cytoplasmic male sterility/fertility restoration system [
9,
10,
11,
12,
13,
14,
15,
16,
17]. Pure line-breeding procedures are also thought to reach the equilibrium point in yield enhancement since they do not produce enough genetic variability. In Indian mustard, hybrids, on the other hand, allow for a greater fraction of genetic variability and a more accessible high heterotic impact [
18]. According to Sodhi et al. [
19], heterosis breeding could be a viable option to pure line breeding for increasing Indian mustard yield potential, as it provides a yield advantage of 19–40% over the best pure line types. As a result, hybrids are one of the most viable alternatives for breaking the yield barriers in Indian mustard.
Indian mustard is exposed to various biotic and abiotic stresses that reduce and limit its output. As a result, in addition to boosting yield potential, the development of stress-tolerant/resistant cultivars is also critical to increase productivity. To achieve sustained and secure yield increase in Indian mustard, plant breeding focuses on crop cultivars with high yield potential and inbuilt resistance to critical yield-limiting factors [
18,
20]. Sclerotinia stem rot, caused by
Sclerotinia sclerotiorum (Lib.) de Bary, is the most destructive fungal disease of Indian mustard at the moment, causing yield losses of 32–90% [
6,
21,
22]. It also impacts the oil content (up to 35%) and quality [
23].
S. sclerotiorum is a cosmopolitan and widespread phytopathogenic fungus with a broad host range that has gone from being inconsequential to symbolic due to global climatic changes and is currently one of the most devastating diseases of Indian mustard. Sclerotia, the survival structure of the pathogen, can survive in plant detritus for many years and act as the major inoculum for infection. It germinates myceliogenically (soil-borne infection) to produce mycelial hyphae that almost instantly invade the lower portion of plants, including the basal stem. In contrast, its carpogenic germination (airborne infection) has apothecia, which are cup-like structures with a 3–6 mm diameter that release ascospores to infect the upper sections of host plants. Almost all plant parts are affected, including cotyledons, leaves, branches, raceme, siliquae, and stems, with infected tissue displaying typical white fluffy cottony mycelial growth symptoms (
Figure 1). On the other hand, infection of the stem causes girdling, which is linked to plant lodging and finally results in significant yield losses in Indian mustard [
1,
6,
24].
Due to its broad infection ability and extended survival ability in the soil, proper treatment of Sclerotinia stem rot using cultural and chemical approaches is challenging, if not impossible. Furthermore, fungicide use is hazardous to the environment and increases the expense of crop production [
6,
25]. One of the most critical aspects of managing Sclerotinia stem rot would be resistant cultivars in Indian mustard [
24]. However, a lack of suitable resistant sources has hampered breeding for resistance in the past. Previous attempts to uncover resistant sources against Sclerotinia stem rot failed miserably because all Indian mustard genotypes tested were sensitive/highly susceptible, and whatever resistant sources were declared were connected to wild and other Brassicaceae species [
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34]. However, in recent years, increased attention has led to identifying a few Indian mustard genotypes resistant to the disease [
1,
6,
33,
34,
35].
Because Sclerotinia stem rot is one of the most critical constraints to Indian mustard production, cultivars with built-in resistance to this disease will be given even more importance to boost and sustain productivity and make this crop more profitable. Next to yield enhancement, breeding for disease resistance is essential in attaining optimal progress in edible oil production to satisfy future demands [
18,
20]. The success of any plant breeding program aimed at incorporating desirable traits, on the other hand, is entirely dependent on the availability of source material and understanding of genetic regulation of the trait(s) in question. As a result, crop breeders are constantly vigilant in determining desirable genetic traits to determine the most practical approach for breeding novel and elite cultivars [
36]. Combining ability analysis frequently aids in selecting the best genotypic combinations for the development of superior hybrids [
37].
Furthermore, plant breeders have a key difficulty in identifying the ideal parental combination to exploit heterosis in the F
1 generation and produce superior transgressive segregants in the F
2 and subsequent segregating generations in any hybridization program. A high
per se performance genotype may not inevitably create better hybrids and/or transgressive segregants when employed in hybridization. Combining ability is a crucial notion that aids in selecting promising parents for hybridization and sheds light on the nature of gene actions that influence superior traits. Line × tester analysis is the most often used of the various mating designs available for combining ability and heterotic effect estimation with knowledge on the genetic control of metric traits in crop plants [
38].
Furthermore, environmental or genotype × environmental interaction may cause variation across genotypes for several traits [
39,
40]. As a result, genotypes should be chosen based on their genetic rather than phenotypic characteristics [
41]. Trait selection necessitates a thorough understanding of the nature and extent of genotypic variation and transmissibility and selection progress. The following selection indicators are commonly used to predict genetic gain under selection: genotypic coefficient of variation(GCV), phenotypic coefficient of variation (PCV), broad-sense heritability (h
2bs), and genetic advance as percent of mean (GAM) [
42,
43]. Furthermore, knowledge of the interrelationship between resistance and yield and its components is critical for determining appropriate selection criteria for breeding Indian mustard for high yield potential and Sclerotinia stem rot resistance. As a result, correlation and path analysis is essential for designing an effective selection strategy and increasing the efficiency of breeding programs [
44,
45]. Plant breeders can use selection indices to fully utilize the response to selection for one or more characteristics. In reality, indices-based selection exhibits the response with direct selection and the correlated response because the selection is competent for other characters. Families from the base population should be assessed to derive genetic and phenotypic estimates such as h
2bs, GAM, genetic correlation, and path coefficients for examining the collection of traits [
46].
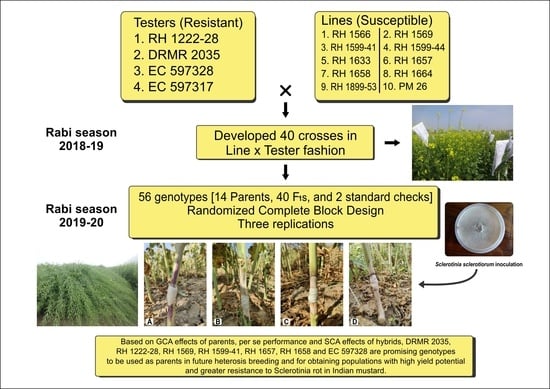
In this context, the current study was carried out to gather information about the nature and extent of gene action, combining ability effects and estimation of selection indices for Sclerotinia stem rot resistance, seed yield, and its component traits in Indian mustard.
4. Discussion
Breeding cultivars with high yield potential and resistance to major diseases is a primary goal of crop improvement projects. When hybridization is attempted in a specific mating design, identifying the best parental genotype combinations allows breeders to take advantage of their heterotic effects and shows that superior transgressive segregants are available in the F
2 and subsequent segregating generations of that cross. The ability to examine the combining ability and heterotic effects in selecting superior parents for the future requires the mean values of parents and F
1 combinations. Line × Tester analysis is the best way to examine the potentiality of contrasting lines (females) and testers (males) for their combining ability and gene action for different traits [
38]. Based on their
per se performance and combining ability effects, the current study was conducted to identify the best parental genotypes and their cross combinations for Sclerotinia stem rot resistance, seed yield, and component traits. The data from the traits analyzed can establish a helpful breeding strategy for future high-yielding Indian mustard hybrids/varieties with increased Sclerotinia stem rot resistance.
There must be sufficient genetic variation within the selected lines for any crop breeding program to succeed. The mean square due to genotypes for agronomic traits, MLL, and DSI was highly significant (
p ≤ 0.01) in the analysis of variance (ANOVA), showing that there was a lot of genotypic variation for these characters among the genotypes tested. As a result, line × tester analysis can split total genetic variance into its appropriate orthogonal components. For most of the traits tested, ANOVA for line × tester mating design demonstrated the significance of their orthogonal components, namely, parents, crosses, and parent vs. Crosses. This implies a high level of genetic variability in both males (testers) and females (lines) parents and their F
1 hybrids, allowing for a more in-depth investigation of genetic variation by combining ability analysis and the measurement of the extent heterosis for these traits. Kaur et al. [
53], Meena et al. [
54,
55,
56], Vaghela et al. [
57], Chaudhary et al. [
58], and Gupta et al. [
9] observed high genetic variability for yield and its component traits in Indian mustard. Godoy et al. [
59], Castano et al. [
60], Achbani et al. [
61], and Grecizes-Besset et al. [
62] in sunflower and Ferreira et al. [
63] in common bean revealed high genetic variability for resistance/susceptibility to
S. sclerotiorum.
Combining ability analysis is frequently used to compare parental performance and better understand the basis of gene action that causes trait manifestation. Furthermore, combining ability is often helpful in forecasting the heterotic response of specific lines/genotypes in various cross combinations and acquiring superior transgressive segregants in the F
2 and subsequent segregating generations. The GCA effect is used to select desirable parents, while the SCA effect is used to evaluate testcross progenies to form heterotic hybrids [
37,
38,
64]. For days to flowering, siliqua length, and 1000-seed weight, significant mean squares attributable to lines and/or testers (GCA) effects and line × tester (SCA) effects show an interplay of additive and non-additive gene effects for the expression of these traits. While additive genetic action influenced the inheritance of days to maturity and plant height, non-additive gene action was significant for expressing the rest of the traits, as revealed by significant mean square due to lines, testers (GCA) and line × tester (SCA) effects for these traits, respectively. Both additive and non-additive genetic effects influenced Sclerotinia stem rot resistance, as evidenced by significant mean squares of lines, testers, and line × tester interactions for both resistance evaluation criteria, namely, MLL and DSI. These findings are consistent with those of Khan et al. [
32], Disi et al. [
30], Godoy et al. [
59], Castano et al. [
60], and Achbani et al. [
61], who found that both additive and non-additive genetic action influenced Sclerotinia rot resistance inheritance and could be improved using the recurrent selection procedure. The significant effect of GCA on the sum of squares of SCA suggested that early generation selection of resistant progenies could be successful.
When utilized in hybridization, selecting parents based on their
per se performance may not always be a fair method because a phenotypically worthy parent may not always produce superior hybrids and transgressive segregants in the segregating generations. As a result, it is necessary to choose parents based on their genetic assets. The parents significant GCA effects are primarily due to their additive and additive × additive gene effects, a fixable component in segregating generations. Based on their GCA effects, parents should be chosen for hybridization to isolate superior segregants in the F
2 and following generations [
38,
63,
64]. Our findings revealed that none of the parents were good general combiners for all of the traits investigated. This conclusion suggests that collective breeding strategies with optimal mating designs must accumulate desirable alleles into a single genetic background. Higher negative GCA values offered better resistance to Sclerotinia stem rot, while higher positive GCA values indicated increased susceptibility. The line RH 1599-41 was involved in three of the top four Sclerotinia stem rot-resistant and early flowering hybrids and had the highest negative GCA effect for DSI and early maturity. Regrettably, it proved to be a poor combiner in seed yield per plant. Positive combining ability and heterotic effects are beneficial for yield component traits like number of primary and secondary branches/plant, seeds/siliqua, siliqua length, and 1000-seed weight because they provide potential for improving yield. Despite ranking second in general effects for yield, the line RH 1569 may be the best choice because it has a good GCA effect for the majority of important yield component traits such as number of primary and secondary branches/plant, number of siliquae on main shoot, and 1000-seed weight, while also having the best general effects for Sclerotinia stem rot resistance. The best general combiners for resistance and the two most crucial yield-related qualities, 1000-seed weight and number of major branches/plant, were DRMR 2035 and RH 1222-28 among testers. Short plant stature and vegetative period are also essential to creating lodging tolerant and comparatively large seed filling period cultivars of Indian mustard for yield and Sclerotinia stem rot resistance. While early maturity allows enough time to raise the following crop, late maturity reduces yield and oil quality due to increased temperature during the final stages of the crop [
65]. Negative combining ability and heterotic effects are thus required for these traits. PM 26 and RH 1658 are good general combiners for shortening vegetative development and reducing plant height.
Overall, the genotypes RH 1569 (line) and DRMR 2035 (tester) looked to be the strongest general combiners for Sclerotinia stem rot resistance and most yield component traits. They should do so well in hybrid combinations with other parents. The greatest criterion for maximizing heterosis in F1 hybrids is to choose parents based on their SCA values. Negative SCA crosses for days to flowering, maturity, and plant height were wanted, while positive SCA crosses for other yield-related attributes were desired.
Contrary to what was expected based on the parent’s GCA, significant SCA effects in the desired direction demonstrate positive deflections with regards to the F
1 crosses. The SCA effect, which considers loci with non-additive and epistatic gene effects, can also identify high heterotic F
1 hybrids. Negative SCA effects for MLL and DSI are desired because they lead to resistance, whereas positive SCA adds to Sclerotinia stem rot susceptibility. RH 1657 × EC 597317 was the only cross that demonstrated a significant desired SCA value for MLL and DSI among other cross combinations, showing the involvement of a particular effect in this hybrid’s resistance expression. It could be because of the good general combiner RH 1657, which has additive effects, and the lousy combiner parent (EC 597317), which has epistatic effects. However, it revealed adverse SCA effects for the majority of yield-related traits. All other crossings, except for cross RH 1657 × EC 597317, had unacceptable and/or insignificant SCA effects for both resistance evaluation criteria, demonstrating that genes/alleles giving Sclerotinia stem rot resistance are recessive over susceptibility. Furthermore, crosses involving both parents with significant GCA effects for resistance had poorer SCA effects, implying the existence of a complex non-allelic gene interaction for resistance and/or that both of these parents may have identical resistance alleles thus could not benefit from fixable gene effects. Similarly, Van Becelaere and Miller [
66] found that GCA effects of both male and female lines were crucial for Sclerotinia head rot resistance in sunflower, but SCA effects were not significant. The ranking of hybrids for resistance assessment parameters in terms of mean values and SCA effects revealed that the lowest mean values did not always predict significant adverse SCA effects, and vice versa. Ross et al. [
67] found a pattern of combining ability effects in grain sorghum, as did Satyanarayana [
68] in rice. Most of the hybrids had insignificant SCA effects for both seed yield and its component traits. The hybrid RH 1658 × EC 597328, on the other hand, had the best SCA effects for yield and components, as well as an insignificant but desirable negative SCA effect for resistance. This cross can develop hybrids or transgressive segregants with excellent seed and oil yields and resistance to Sclerotinia stem rot. The cross RH 1599-44 × EC 597317 showed significant SCA effects for lowering plant height and vegetative and maturity periods in this study. As a result, including the parents in a specific mating design such as diallel or triallel may increase the possibility of producing high-yielding, resistant segregants and developing hybrids. The inconsistency of GCA and SCA effects suggests that these traits have complex gene connections.
The contribution to the total variation of the lines, testers, and their interactions support prior results that general effects were more relevant than specific effects for Sclerotinia rot resistance [
59,
61,
62]. On the other hand, specific effects had a greater impact on yield component traits and oil content [
9,
11,
54,
57]. GCA and SCA variances revealed the role of both additive and non-additive gene action in the expression of most examined traits. The significance of SCA in creating heterotic crosses for most yield-related traits was highlighted by higher SCA variance than GCA variance of lines and testers. The allele frequencies between parental genotypes determine the magnitude of the GCA/SCA variance ratio. For most of the yield-attributing traits, such as number of primary and secondary branches/plant, main shoot length, number of siliquae on main shoot, siliqua length, number of seeds/siliqua, and oil content, this ratio revealed the predominance of non-additive gene action (SCA variance). This indicates that selecting superior plants for these traits should be deferred to subsequent generations. Although additive gene action (GCA variance) was essential for days to flowering, days to maturity, plant height, and Sclerotinia rot resistance, selection of phenotypically superior plants would be effective in early generations. These findings are consistent with those of Meena et al. [
54], Vaghela et al. [
57], and Gupta et al. [
69], but differ from those of Dahiya et al. [
70] and Chaudhary et al. [
58]. Because both additive and non-additive gene actions were necessary for the inheritance of the studied traits, hybridization methods that simultaneously use additive and non-additive gene effects, such as diallel selective mating scheme or reciprocal recurrent crossing, could be helpful in the genetic improvement of the traits under consideration.
The current study’s PCV values were larger than the GCV values for all traits, showing that environmental effects on the characteristics were more relevant than genotypic effects. Previous publications on higher PCV values than GCV values for yield and component traits in Indian mustard [
71,
72,
73,
74,
75] complement our findings. Seed yield/plant had a considerable discrepancy for PCV and GCV, owing to the greater confounding effect of environment on this characteristic. Furthermore, due to the concealing effect of the environment, GCV and PCV estimates do not substantiate the exhaustive extent of heritable variation, which may be assessed more precisely with heritability and genetic advance estimates [
76]. Heritability and genetic advancement of a particular trait determine genetic gain or responsiveness to selection. Singh [
77] divided broad-sense heritability estimates (h
2bs) into four categories: low (<40%), moderate (40–59%), moderately high (60–79%), and extremely high (>80%). Days to 50% flowering, 1000-seed weight, oil content, number of primary and secondary branches/plant, main shoot length, number of siliquae on main shoot, siliqua length, MLL, and DSI all had moderately high to extremely high heritability values in this study. If these traits are employed to boost seed and oil yield and Sclerotinia stems rot resistance in Indian mustard, the projected gain from the selection will be considerable. The high heritability estimates also suggest that the environment poorly influences certain traits. Heritability estimates for plant height and number of seeds/siliqua were intermediate, indicating that these parameters’ environmental and genetic effects were indistinguishable. Days to maturity and seed yield/plant have low heritability estimates, and direct selection for these traits would be challenging due to the high environmental impact. As a result, component traits with high heritability and strong desirable correlations can be picked concurrently to select these traits. Several studies found moderate to high heritability for yield and its component traits in Indian mustard, similar to the current study [
78,
79,
80,
81,
82,
83]. The heritability of Sclerotinia stem rot resistance evaluation parameters (MLL and DSI) was moderate, indicating that early selection for Sclerotinia stem rot resistance may be successful. Sclerotinia stem rot resistance has been estimated to be heritable by various researchers, with values ranging from 21 to 88% [
32,
84,
85]. Genetic advance as percent of mean (GAM) values are divided into three categories: low (0–10%), moderate (10–20%), and high (>20%) [
86]. High GAM values for DSI, MLL, 1000-seed weight, and days to 50% flowering imply that these traits are highly likely to be improved through selection. The rest of the traits have intermediate to low GAM values, indicating that selection in the early generation is ineffective in improving these traits. High heritability combined with moderate to high GAM indicates that component traits are primarily driven by genes and are only marginally influenced by the environment, making them easily exploitable for seed yield improvement [
87]. Selection is difficult for a character with low to moderate h
2bs and low GAM [
88]. Selection for high-yielding genotypes with inbuilt resistance to Sclerotinia stem rot should be delayed to confirm the homozygosity of genes controlling resistance and yield and its component traits. It should be based on a high number of primary and secondary branches/plant, longer main shoot length, high number of siliquae on main shoot, more 1000-seed weight, fewer days to 50% flowering, and low MLL and DSI under multiple locations and years.
For most of the traits tested, the genotypic correlation was higher than the phenotypic correlation, showing genetic relationships. As a result, phenotype-based selection would work [
82,
89,
90]. Seed yield in mustard is determined by several interdependent and environment-dependent traits. The number of primary and secondary branches, main shoot length, number of siliquae on main shoot, siliqua length, number of seeds/siliqua, and 1000-seed weight all had positive and significant genotypic relationships in the current study. This shows that seed yield will be improved by indirect selection through component traits. Similarly, in Indian mustard [
78,
91,
92,
93,
94] a strong link between these characteristics and seed yield has been found. In addition, days to flowering showed a significant and negative relationship with seed yield, which is beneficial because early flowering in Indian mustard allowed for shorter vegetative and reproductive stages and increased grain filling time. However, because the development of high-yielding and early maturing varieties is the primary goal of mustard breeders, the significant and positive correlation between seed yield/plant and days to maturity is undesired. Indirect selection of yield through its component traits necessitates the examination of genotypic and phenotypic relationships among the traits. However, the correlation-based choice is ineffective because it only shows the linear relationship between two traits.
Path coefficient analysis has been widely used in crop breeding projects to discover the relationship between seed yield and component traits [
87]. Days to 50% flowering, 1000-seed weight, number of primary branches/plant, and main shoot length exhibited very high and positive direct effects on seed yield/plant in this study, indicating that these traits should be prioritized for indirect selection. However, through the MLL and DSI, siliqua length negatively impacted seed yield. To generate high-yielding and disease-resistant cultivars, extensive genetic analysis and identification of alleles causing linkage drag among Sclerotinia stem rot resistance and seed yield are necessary. Through siliqua length, 1000-seed weight had the greatest indirect effect on seed yield, implying that longer siliquae are more likely to have higher seed weight. Our findings are supported by Lodhi et al. [
95], who used path analysis to examine 90 different Indian mustard genotypes and found that main shoot length, number of primary branches/plant, and number of seeds/siliqua had the most significant direct effect on seed yield. Pandey et al. [
96] found that plant height, 1000-seed weight, number of seed/siliqua, siliqua on main raceme, and primary branches/plant had the greatest direct effect on seed yield. Saroj et al. [
83] found that total seed yield had the highest positive direct effect, followed by siliquae on the main shoot and seed size, in a separate study involving 289 diverse Indian mustard accessions analyzed over two seasons.