Nutritional Value of Eggplant Cultivars and Association with Sequence Variation in Genes Coding for Major Phenolics
Abstract
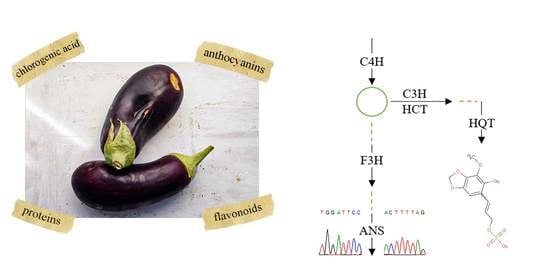
:1. Introduction
2. Results–Discussion
2.1. Fruit Quality Characteristics
2.2. Exon Sequences and Translation
2.3. Gene Mutations, SNPs, and Metabolites Associations
3. Materials and Methods
3.1. Plant Material
3.2. Chemical Analysis
3.2.1. Fruit Extraction
3.2.2. Colorimetric Determination of Total Phenolics, Total Monomeric Anthocyanins, Total Flavonoids, Proteins, and Sugars
3.2.3. Quantification of Chlorogenic Acid Content by Using an HPLC-UV Method
3.2.4. Colorimetric Determination of Antioxidant Capacity (DPPH and FRAP)
3.3. Molecular Analysis
3.3.1. DNA Extraction and PCR Amplification
3.3.2. Molecular Data Analysis
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gramazio, P.; Chatziefstratiou, E.; Petropoulos, C.; Chioti, V.; Mylona, P.; Kapotis, G.; Vilanova, S.; Prohens, J.; Papasotiropoulos, V. Multi-Level Characterization of Eggplant Accessions from Greek Islands and the Mainland Contributes to the Enhancement and Conservation of this Germplasm and Reveals a Large Diversity and Signatures of Differentiation between both Origins. Agronomy 2019, 9, 887. [Google Scholar] [CrossRef]
- Raigón, M.D.; Prohens, J.; Muñoz-Falcón, J.E.; Nuez, F. Comparison of eggplant landraces and commercial varieties for fruit content of phenolics, minerals, dry matter and protein. J. Food Compos. Anal. 2008, 21, 370–376. [Google Scholar] [CrossRef]
- Stommel, J.R.; Whitaker, B.D. Phenolic Acid Content and Composition of Eggplant Fruit in a Germplasm Core Subset. J. Am. Soc. Hortic. Sci. 2003, 128, 704–710. [Google Scholar] [CrossRef]
- Cao, G.; Sofic, E.; Prior, R.L. Antioxidant Capacity of Tea and Common Vegetables. J. Agric. Food Chem. 1996, 44, 3426–3431. [Google Scholar] [CrossRef]
- Whitaker, A.B.D.; Stommel, J.R. Distribution of Hydroxycinnamic Acid Conjugates in Fruit of Commercial Eggplant (Solanum melongena L.) Cultivars. J. Agric. Food Chem. 2003, 51, 3448–3454. [Google Scholar] [CrossRef]
- Niño-Medina, G.; Urías-Orona, V.; Rangel, M.D.M.; Heredia, J.B. Structure and content of phenolics in eggplant (Solanum melongena)—A review. S. Afr. J. Bot. 2017, 111, 161–169. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Plazas, M.; Andújar, I.; Vilanova, S.; Hurtado, M.; Gramazio, P.; Herraiz, F.J.; Prohens, J. Breeding for Chlorogenic Acid Content in Eggplant: Interest and Prospects. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 26. [Google Scholar] [CrossRef]
- Gürbüz, N.; Uluişik, S.; Frary, A.; Frary, A.; Doğanlar, S. Health benefits and bioactive compounds of eggplant. Food Chem. 2018, 268, 602–610. [Google Scholar] [CrossRef]
- Prohens, J.; Muñoz, J.E.; Vilanova, S.; Castro, Á.; Ribas, F.; Nuez, F. Participatory breeding in eggplant: Selection and improvement for quality and yield in a local landrace. In Progress in Research on Capsicum & Eggplant; Niemirowicz-Szczytt, K., Ed.; Warsaw University of Life Sciences Press: Warsaw, Poland, 2007; pp. 221–230. [Google Scholar]
- Singh, A.P.; Luthria, D.; Wilson, T.; Vorsa, N.; Singh, V.; Banuelos, G.S.; Pasakdee, S. Polyphenols content and antioxidant capacity of eggplant pulp. Food Chem. 2009, 114, 955–961. [Google Scholar] [CrossRef]
- Hanson, P.M.; Yang, R.-Y.; Tsou, S.C.; Ledesma, D.; Engle, L.; Lee, T.-C. Diversity in eggplant (Solanum melongena) for superoxide scavenging activity, total phenolics, and ascorbic acid. J. Food Compos. Anal. 2006, 19, 594–600. [Google Scholar] [CrossRef]
- Zambrano-Moreno, E.L.; Chávez-Jáuregui, R.N.; Plaza, M.D.L.; Wessel-Beaver, L. Phenolic content and antioxidant capacity in organically and conventionally grown eggplant (Solanum melongena) fruits following thermal processing. Food Sci. Technol. 2015, 35, 414–420. [Google Scholar] [CrossRef]
- Kaushik, P.; Andújar, I.; Vilanova, S.; Plazas, M.; Gramazio, P.; Herraiz, F.J.; Brar, N.S.; Prohens, J. Breeding Vegetables with Increased Content in Bioactive Phenolic Acids. Molecules 2015, 20, 18464–18481. [Google Scholar] [CrossRef]
- Prohens, J.; Whitaker, B.; Plazas, M.; Vilanova, S.; Hurtado, M.; Blasco, M.; Gramazio, P.; Stommel, J. Genetic diversity in morphological characters and phenolic acids content resulting from an interspecific cross between eggplant, Solanum melongena, and its wild ancestor (S. incanum). Ann. Appl. Biol. 2013, 162, 242–257. [Google Scholar] [CrossRef]
- Plazas, M.; Prohens, J.; Cuñat, A.N.; Vilanova, S.; Gramazio, P.; Herraiz, F.J.; Andújar, I. Reducing Capacity, Chlorogenic Acid Content and Biological Activity in a Collection of Scarlet (Solanum aethiopicum) and Gboma (S. macrocarpon) Eggplants. Int. J. Mol. Sci. 2014, 15, 17221–17241. [Google Scholar] [CrossRef]
- Kaushik, P.; Gramazio, P.; Vilanova, S.; Raigón, M.D.; Prohens, J.; Plazas, M. Phenolics content, fruit flesh colour and browning in cultivated eggplant, wild relatives and interspecific hybrids and implications for fruit quality breeding. Food Res. Int. 2017, 102, 392–401. [Google Scholar] [CrossRef]
- Kaushik, P. Line × Tester Analysis for Morphological and Fruit Biochemical Traits in Eggplant (Solanum melongena L.) Using Wild Relatives as Testers. Agronomy 2019, 9, 185. [Google Scholar] [CrossRef]
- Clifford, M.N.; Jaganath, I.B.; Ludwig, I.A.; Crozier, A. Chlorogenic acids and the acyl-quinic acids: Discovery, biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017, 34, 1391–1421. [Google Scholar] [CrossRef]
- Sękara, A.; Cebula, S.; Kunicki, E. Cultivated eggplants—Origin, breeding objectives and genetic resources, a review. World 2007, 19, 97–114. [Google Scholar]
- Zhang, Y.; Chu, G.; Hu, Z.; Gao, Q.; Cui, B.; Tian, S.; Wang, B.; Chen, G. Genetically engineered anthocyanin pathway for high health-promoting pigment production in eggplant. Mol. Breed. 2016, 36, 1–14. [Google Scholar] [CrossRef]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S.; et al. Draft Genome Sequence of Eggplant (Solanum melongena L.): The Representative Solanum Species Indigenous to the Old World. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef]
- Li, D.; Qian, J.; Li, W.; Jiang, Y.; Gan, G.; Li, W.; Chen, R.; Yu, N.; Li, Y.; Wu, Y. Genome sequence and analysis of the eggplant (Solanum melongena L.). BioRxiv 2019, 824540. Available online: https://www.biorxiv.org/content/10.1101/824540v1 (accessed on 12 May 2022).
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2016, 6, 1233. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, V.G.; Subbotina, N.M.; Maluchenko, O.P.; Lebedeva, T.N.; Krutovsky, K.V.; Shestibratov, K.A. Transferability and Polymorphism of SSR Markers Located in Flavonoid Pathway Genes in Fragaria and Rubus Species. Genes 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Okmen, B.; Sigva, H.O.; Mutlu, S.; Doganlar, S.; Yemenicioglu, A.; Frary, A. Total Antioxidant Activity and Total Phenolic Contents in Different Turkish Eggplant (Solanum melongena L.) Cultivars. Int. J. Food Prop. 2009, 12, 616–624. [Google Scholar] [CrossRef]
- Martínez-Ispizua, E.; Calatayud, Á.; Marsal, J.I.; Mateos-Fernández, R.; Díez, M.J.; Soler, S.; Valcárcel, J.V.; Martínez-Cuenca, M.-R. Phenotyping Local Eggplant Varieties: Commitment to Biodiversity and Nutritional Quality Preservation. Front. Plant Sci. 2021, 12, 696272. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, M.; Xiao, Y.; Cui, D.; Qin, Y.; Wu, J.; Wang, W.; Wang, G. Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L. Int. J. Mol. Sci. 2018, 19, 789. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin Accumulation and Molecular Analysis of Anthocyanin Biosynthesis-Associated Genes in Eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Stout, J.; Weng, J.-K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef]
- Sanath Hettiarachi, S.H.; Hettiarachchi, P.L. Biotechnologies for increasing Antioxidant Production from Plants. In Plants as a Source of Natural Antioxidants; Dubey, N.K., Ed.; CABI: Wallingford, UK, 2014; pp. 156–168. [Google Scholar]
- Page, M.; Sultana, N.; Paszkiewicz, K.; Florance, H.; Smirnoff, N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2012, 35, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laurentin, A.; Edwards, C.A. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 2003, 315, 143–145. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic– phosphotungstic acid reagent. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Woisky, R.G.; Salatino, A. Analysis of propolis: Some parameters and procedures for chemical quality control. J. Apic. Res. 1998, 37, 99–105. [Google Scholar] [CrossRef]
- Giusti, M.M.; Wrolstad, R.E. Characterization and Measurement of Anthocyanins by UV-visible Spectroscopy. Handbook Food Anal, Chem. 2001, 2, 19–31. [Google Scholar] [CrossRef]
- Lee, J.; Rennaker, C.; Wrolstad, R. Comparison of two methods for anthocyanin quantification. Acta Hortic. 2009, 831–834. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Huang, D.; Ou, B.; Prior, R.L. The Chemistry behind Antioxidant Capacity Assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Diamanti, J.; Battino, M.; Mezzetti, B. Breeding for fruit nutritional and nutraceutical quality. Book Chapter. In Breeding for Fruit Quality; Jenks, M.A., Bebeli, P.J., Eds.; Wiley-Blackwell: Chichester, UK, 2011; pp. 61–79. [Google Scholar] [CrossRef]

| Cultivar | Origin | Fruit Color | Shape | Fruit Weight (g) |
|---|---|---|---|---|
| ‘Lagkada’ (n = 3) | GR | Deep purple | Cylindrical | 278.3 ± 39.6 (22.8) |
| ‘Tsakoniki’ (n = 3) | GR | Purple with white stripes | Cylindrical | 194.3 ± 19.1 (11.1) |
| ‘ΕΜΙ’ (n = 3) | GR | Deep purple | Oval | 481.7 ± 51.3 (29.6) |
| ‘Cristal F1’ (n = 6) | ES | Deep purple | Oval | 309.2 ± 31.9 (10.6) |
| ‘Sabelle F1’ (n = 3) | NL | Light purple | Round | 349.7 ± 58.4 (33.7) |
| ‘Angela F1’ (n = 3) | NL | Purple with white stripes | Oval | 236.7 ± 2.1 (1.2) |
| ‘Nilo F1’ (n = 3) | NL | Deep purple | Cylindrical | 122.7 ± 6.7 (3.8) |
| ‘Leticia F1’ (n = 9) | NL | Deep purple | Oval | 332.5 ± 48.7 (16.2) |
| ‘Monarca F1’ (n = 3) | NL | Deep purple | Oval | 262.7 ± 22.7 (13.1) |
| ‘Lydia’ F1’ (n = 9) | NL | Purple with white stripes | Oval | 270.2 ± 36.7 (15.0) |
| ‘Samantha F1 (n = 3) | NL | Light green | Oval | 304.7 ± 70.4 (40.7) |
| ‘Lato F1’ (n = 6) | NL | White | Oval | 254.7 ± 10.4 (4.2) |
| ‘Blanchette F1’ (n = 3) | NL | White | Round | 196.7 ± 17.6 (10.1) |
| Weight | TPC | TFC | TMA | AsA | Proteins | Sugars | CQA | FRAP | DPPH | |
|---|---|---|---|---|---|---|---|---|---|---|
| Weight | - | |||||||||
| TPC | 0.266 * | - | ||||||||
| TFC | 0.097 | 0.448 *** | - | |||||||
| TMA | 0.456 *** | 0.689 *** | 0.167 | - | ||||||
| AsA | 0.508 *** | 0.502 *** | 0.393 ** | 0.690 *** | - | |||||
| Proteins | 0.231 | 0.514 *** | -0.180 | 0.619 *** | 0.405 ** | - | ||||
| Sugars | 0.002 | 0.367 ** | 0.128 | 0.423 ** | 0.271 * | 0.392 ** | - | |||
| CQA | −0.156 | 0.327 * | 0.288 * | 0.321 * | 0.142 | 0.184 | 0.068 | - | ||
| FRAP | −0.033 | 0.422 ** | 0.119 | 0.366 ** | 0.290 * | 0.134 | 0.107 | 0.400 * | - | |
| DPPH | 0.417 ** | 0.761 *** | 0.337 * | 0.898 *** | 0.703 *** | 0.560 *** | 0.307 * | 0.379 ** | 0.363 ** | - |
| Gene/Exon | Alignment Position | Type of Mutation | Substitution | Translation |
|---|---|---|---|---|
| C4H4 | 108 | missense | A>G | Thr>Ala |
| 413 | synonymous | T>C | Ile | |
| HQT | 105 | synonymous | T>C | His |
| 438 | missense | T>G | His>Gln | |
| 565 | missense | A>G | Thr>Ala | |
| 996 | synonymous | T>C | Ser | |
| F3H | 163 | missense | A>G | Lys>Glu |
| ANS1 | 65 | missense | T>C | Val>Ala |
| 141 | missense | T>A | Asp>Glu | |
| 186 | synonymous | A>G | Ser | |
| 282 | synonymous | T>C | His | |
| MYB1 | 52 | missense | T>A | Trp>Arg |
| 675 | synonymous | C>A | Glu | |
| 678 | synonymous | C>A | Ala | |
| 737 | missense | T>C | Phe>Ser | |
| 760 | synonymous | T>C | Leu |
| C4H4 | HQT | F3H | ANS1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 108 | 413 | 105 | 438 | 565 | 996 | 163 | 65 | 141 | 186 | 282 | |
| TPC | 0.109 | 0.109 | 0.060 | 0.460 | 0.460 | 0.460 | 0.460 | 0.143 | 0.143 | 0.143 | 0.143 |
| TFC | 0.022 | 0.022 | 0.877 | 0.004 | 0.004 | 0.004 | 0.064 | 0.000 | 0.000 | 0.000 | 0.000 |
| TMA | 0.000 | 0.000 | 0.000 | 0.050 | 0.050 | 0.050 | 0.050 | 0.001 | 0.001 | 0.001 | 0.001 |
| AsA | 0.087 | 0.087 | 0.029 | 0.130 | 0.130 | 0.130 | 0.068 | 0.098 | 0.098 | 0.098 | 0.098 |
| Proteins | 0.027 | 0.027 | 0.000 | 0.050 | 0.050 | 0.050 | 0.050 | 0.014 | 0.014 | 0.014 | 0.014 |
| Sugars | 0.145 | 0.145 | 0.077 | 0.494 | 0.494 | 0.494 | 0.494 | 0.041 | 0.041 | 0.041 | 0.041 |
| CQA | 0.632 | 0.632 | 0.183 | 0.008 | 0.008 | 0.008 | 0.347 | 0.526 | 0.526 | 0.526 | 0.526 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chioti, V.; Zeliou, K.; Bakogianni, A.; Papaioannou, C.; Biskinis, A.; Petropoulos, C.; Lamari, F.N.; Papasotiropoulos, V. Nutritional Value of Eggplant Cultivars and Association with Sequence Variation in Genes Coding for Major Phenolics. Plants 2022, 11, 2267. https://doi.org/10.3390/plants11172267
Chioti V, Zeliou K, Bakogianni A, Papaioannou C, Biskinis A, Petropoulos C, Lamari FN, Papasotiropoulos V. Nutritional Value of Eggplant Cultivars and Association with Sequence Variation in Genes Coding for Major Phenolics. Plants. 2022; 11(17):2267. https://doi.org/10.3390/plants11172267
Chicago/Turabian StyleChioti, Vasileia, Konstantina Zeliou, Aikaterini Bakogianni, Charikleia Papaioannou, Antonis Biskinis, Constantinos Petropoulos, Fotini N. Lamari, and Vasileios Papasotiropoulos. 2022. "Nutritional Value of Eggplant Cultivars and Association with Sequence Variation in Genes Coding for Major Phenolics" Plants 11, no. 17: 2267. https://doi.org/10.3390/plants11172267