The Modification of Plant Cell Wall Polysaccharides in Potato Plants during Pectobacterium atrosepticum-Caused Infection
Abstract
:1. Introduction
2. Results
2.1. Biochemical Analysis of PCW Polysaccharides during Pba-Caused Infection
2.1.1. Polysaccharides of Buffer-Extractable Fraction
2.1.2. Polysaccharides of the Ammonium Oxalate Extractable Fraction
2.1.3. Polysaccharides of KOH-Extractable Fraction
2.2. Immunolocalisation of PCW Polysaccharides and Infection-Related Modification of Different Plant Tissues
2.3. Plant and Bacteria PCW-Related Gene Expression
2.3.1. Plant Genes Related to CLG Metabolism
2.3.2. Plant Genes Related to Pectin Metabolism
2.3.3. Plant Genes Encoding Other Cell Wall Proteins
2.3.4. Pba Genes Encoding Plant Cell Wall Degrading Enzymes (PCWDEs)
3. Discussion
3.1. The Modification of Pectic Compounds in Potato Plants during Pba-Caused Infection
3.2. The Modification of CLGs in Potato Plants during Pba-Caused Infection
4. Materials and Methods
4.1. Growth Conditions for Plants and Bacteria, Plant Inoculation, and Sample Collection
4.2. RNA Extraction and cDNA Library Preparation and Sequencing
4.3. RNA-Seq Data Processing and Analysis of Differential Expression
4.4. Phylogenetic Analysis
4.5. Plant Cell Wall (PCW) Isolation and Fractionation
4.6. Molecular Weight Distribution Analysis
4.7. Monosaccharide Analysis
4.8. Microscopy and Immunocytochemistry
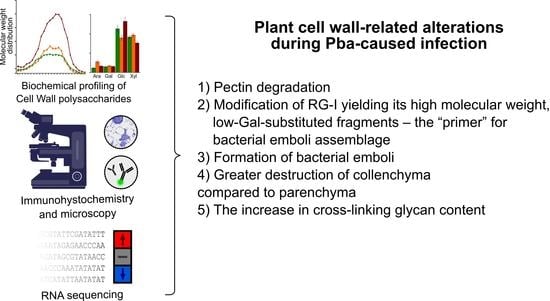
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahal, D.; Pich, A.; Braun, H.P.; Wydra, K. Analysis of Cell Wall Proteins Regulated in Stem of Susceptible and Resistant Tomato Species after Inoculation with Ralstonia solanacearum: A Proteomic Approach. Plant Mol. Biol. 2010, 73, 643–658. [Google Scholar] [CrossRef] [Green Version]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The Role of the Cell Wall in Plant Immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The Role of the Secondary Cell Wall in Plant Resistance to Pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Cantu, D.; Vicente, A.R.; Greve, L.C.; Dewey, F.M.; Bennett, A.B.; Labavitch, J.M.; Powell, A.L.T. The Intersection between Cell Wall Disassembly, Ripening, and Fruit Susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 2008, 105, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Sharmin, S.; Azam, M.S.; Islam, M.S.; Sajib, A.A.; Mahmood, N.; Hasan, A.M.M.; Ahmed, R.; Sultana, K.; Khan, H. Xyloglucan Endotransglycosylase/Hydrolase Genes from a Susceptible and Resistant Jute Species Show Opposite Expression Pattern Following Macrophomina phaseolina Infection. Commun. Integr. Biol. 2012, 5, 598–606. [Google Scholar] [CrossRef]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant Cell Wall Dynamics and Wall-Related Susceptibility in Plant–Pathogen Interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Blanco, C.; Feng, D.X.; Hu, J.; Sánchez-Vallet, A.; Deslandes, L.; Llorente, F.; Berrocal-Lobo, M.; Keller, H.; Barlet, X.; Sánchez-Rodríguez, C.; et al. Impairment of Cellulose Synthases Required for Arabidopsis Secondary Cell Wall Formation Enhances Disease Resistance. Plant Cell 2007, 19, 890–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manabe, Y.; Nafisi, M.; Verhertbruggen, Y.; Orfila, C.; Gille, S.; Rautengarten, C.; Cherk, C.; Marcus, S.E.; Somerville, S.; Pauly, M.; et al. Loss-of-Function Mutation of Reduced Wall Acetylation2 in Arabidopsis Leads to Reduced Cell Wall Acetylation and Increased Resistance to Botrytis cinerea. Plant Physiol. 2011, 155, 1068–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, A.; Miedes, E.; Bacete, L.; Rodríguez, T.; Mélida, H.; Denancé, N.; Sánchez-Vallet, A.; Rivière, M.-P.; López, G.; Freydier, A.; et al. Arabidopsis Cell Wall Composition Determines Disease Resistance Specificity and Fitness. Proc. Natl. Acad. Sci. USA 2021, 118, e2010243118. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, V.Y.; Daminova, A.G.; Mikshina, P.V.; Petrova, O.E.; Ageeva, M.V.; Salnikov, V.V.; Gorshkova, T.A.; Gogolev, Y.V. Pathogen-Induced Conditioning of the Primary Xylem Vessels—A Prerequisite for the Formation of Bacterial Emboli by Pectobacterium atrosepticum. Plant Biol. 2016, 18, 609–617. [Google Scholar] [CrossRef]
- Gijsegem, F.V.; van der Wolf, J.M.; Toth, I. Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Cham, Switzerland, 2021; ISBN 978-3-030-61458-4. [Google Scholar] [CrossRef]
- Gorshkov, V.; Daminova, A.; Ageeva, M.; Petrova, O.; Gogoleva, N.; Tarasova, N.; Gogolev, Y. Dissociation of a Population of Pectobacterium atrosepticum SCRI1043 in Tobacco Plants: Formation of Bacterial Emboli and Dormant Cells. Protoplasma 2014, 251, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Tsers, I.; Gorshkov, V.; Gogoleva, N.; Parfirova, O.; Petrova, O.; Gogolev, Y. Plant Soft Rot Development and Regulation from the Viewpoint of Transcriptomic Profiling. Plants 2020, 9, 1176. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving Views of Pectin Biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liepman, A.H.; Nairn, C.J.; Willats, W.G.T.; Sørensen, I.; Roberts, A.W.; Keegstra, K. Functional Genomic Analysis Supports Conservation of Function among Cellulose Synthase-like a Gene Family Members and Suggests Diverse Roles of Mannans in Plants. Plant Physiol. 2007, 143, 1881–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-J.; Chandrasekar, B.; Rea, A.C.; Danhof, L.; Zemelis-Durfee, S.; Thrower, N.; Shepard, Z.S.; Pauly, M.; Brandizzi, F.; Keegstra, K. The Synthesis of Xyloglucan, an Abundant Plant Cell Wall Polysaccharide, Requires CSLC Function. Proc. Natl. Acad. Sci. USA 2020, 117, 20316–20324. [Google Scholar] [CrossRef]
- Yin, Y.; Johns, M.A.; Cao, H.; Rupani, M. A Survey of Plant and Algal Genomes and Transcriptomes Reveals New Insights into the Evolution and Function of the Cellulose Synthase Superfamily. BMC Genomics 2014, 15, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madson, M.; Dunand, C.; Li, X.; Verma, R.; Vanzin, G.F.; Caplan, J.; Shoue, D.A.; Carpita, N.C.; Reiter, W.-D. The MUR3 Gene of Arabidopsis Encodes a Xyloglucan Galactosyltransferase That Is Evolutionarily Related to Animal Exostosins. Plant Cell 2003, 15, 1662–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Peña, M.J.; Zhou, G.-K.; Nairn, C.J.; Wood-Jones, A.; Richardson, E.A.; Morrison, W.H.; Darvill, A.G.; York, W.S.; Ye, Z.-H. Arabidopsis Fragile Fiber8, Which Encodes a Putative Glucuronyltransferase, Is Essential for Normal Secondary Wall Synthesis. Plant Cell 2005, 17, 3390–3408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, A.-M.; Rihouey, C.; Seveno, M.; Hörnblad, E.; Singh, S.K.; Matsunaga, T.; Ishii, T.; Lerouge, P.; Marchant, A. The Arabidopsis IRX10 and IRX10-LIKE Glycosyltransferases Are Critical for Glucuronoxylan Biosynthesis during Secondary Cell Wall Formation. Plant J. 2009, 57, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.K.; Schultink, A.; Keegstra, K.; Wilkerson, C.G.; Pauly, M. RNA-Seq Analysis of Developing Nasturtium Seeds (Tropaeolum majus): Identification and Characterization of an Additional Galactosyltransferase Involved in Xyloglucan Biosynthesis. Mol. Plant 2012, 5, 984–992. [Google Scholar] [CrossRef] [Green Version]
- Harholt, J.; Jensen, J.K.; Verhertbruggen, Y.; Søgaard, C.; Bernard, S.; Nafisi, M.; Poulsen, C.P.; Geshi, N.; Sakuragi, Y.; Driouich, A.; et al. ARAD Proteins Associated with Pectic Arabinan Biosynthesis Form Complexes When Transiently Overexpressed in planta. Planta 2012, 236, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.-M.; Hörnblad, E.; Voxeur, A.; Gerber, L.; Rihouey, C.; Lerouge, P.; Marchant, A. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L Pairs of Glycosyltransferase Genes Reveals Critical Contributions to Biosynthesis of the Hemicellulose Glucuronoxylan. Plant Physiol. 2010, 153, 542–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voiniciuc, C.; Günl, M.; Schmidt, M.H.-W.; Usadel, B. Highly Branched Xylan Made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 Links Mucilage to Arabidopsis Seeds. Plant Physiol. 2015, 169, 2481–2495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gille, S.; de Souza, A.; Xiong, G.; Benz, M.; Cheng, K.; Schultink, A.; Reca, I.-B.; Pauly, M. O-Acetylation of Arabidopsis Hemicellulose Xyloglucan Requires AXY4 or AXY4L, Proteins with a TBL and DUF231 Domain. Plant Cell 2011, 23, 4041–4053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Cui, D.; Ye, Z.-H. Regiospecific Acetylation of Xylan Is Mediated by a Group of DUF231-Containing O-Acetyltransferases. Plant Cell Physiol. 2017, 58, 2126–2138. [Google Scholar] [CrossRef] [Green Version]
- Stranne, M.; Ren, Y.; Fimognari, L.; Birdseye, D.; Yan, J.; Bardor, M.; Mollet, J.-C.; Komatsu, T.; Kikuchi, J.; Scheller, H.V.; et al. TBL10 Is Required for O-Acetylation of Pectic Rhamnogalacturonan-I in Arabidopsis thaliana. Plant J. 2018, 96, 772–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takenaka, Y.; Kato, K.; Ogawa-Ohnishi, M.; Tsuruhama, K.; Kajiura, H.; Yagyu, K.; Takeda, A.; Takeda, Y.; Kunieda, T.; Hara-Nishimura, I.; et al. Pectin RG-I Rhamnosyltransferases Represent a Novel Plant-Specific Glycosyltransferase Family. Nat. Plants 2018, 4, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Tovar-Herrera, O.E.; Rodríguez, M.; Olarte-Lozano, M.; Sampedro-Guerrero, J.A.; Guerrero, A.; Pinto-Cámara, R.; Alvarado-Affantranger, X.; Wood, C.D.; Moran-Mirabal, J.M.; Pastor, N.; et al. Analysis of the Binding of Expansin Exl1, from Pectobacterium carotovorum, to Plant Xylem and Comparison to EXLX1 from Bacillus subtilis. ACS Omega 2018, 3, 7008–7018. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.; Blanco, C.; Condemine, G.; Expert, D.; Franza, T.; Hayes, C.; Hugouvieux-Cotte-Pattat, N.; López Solanilla, E.; Low, D.; Moleleki, L.; et al. The Role of Secretion Systems and Small Molecules in Soft-Rot Enterobacteriaceae Pathogenicity. Annu. Rev. Phytopathol. 2012, 50, 425–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarasova, N.; Gorshkov, V.; Petrova, O.; Gogolev, Y. Potato Signal Molecules That Activate Pectate Lyase Synthesis in Pectobacterium atrosepticum SCRI1043. World J. Microbiol. Biotechnol. 2013, 29, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial Pectate Lyases, Structural and Functional Diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, V.; Gubaev, R.; Petrova, O.; Daminova, A.; Gogoleva, N.; Ageeva, M.; Parfirova, O.; Prokchorchik, M.; Nikolaichik, Y.; Gogolev, Y. Transcriptome Profiling Helps to Identify Potential and True Molecular Switches of Stealth to Brute Force Behavior in Pectobacterium atrosepticum during Systemic Colonization of Tobacco Plants. Eur. J. Plant Pathol. 2018, 152, 957–976. [Google Scholar] [CrossRef]
- Gorshkov, V.; Islamov, B.; Mikshina, P.; Petrova, O.; Burygin, G.; Sigida, E.; Shashkov, A.; Daminova, A.; Ageeva, M.; Idiyatullin, B.; et al. Pectobacterium atrosepticum Exopolysaccharides: Identification, Molecular Structure, Formation under Stress and in Planta Conditions. Glycobiology 2017, 27, 1016–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otulak-Kozieł, K.; Kozieł, E.; Bujarski, J.J. Spatiotemporal Changes in Xylan-1/Xyloglucan and Xyloglucan Xyloglucosyl Transferase (XTH-Xet5) as a Step-In of Ultrastructural Cell Wall Remodelling in Potato–Potato Virus Y (PVYNTN) Hypersensitive and Susceptible Reaction. Int. J. Mol. Sci. 2018, 19, 2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCartney, L.; Marcus, S.E.; Knox, J.P. Monoclonal Antibodies to Plant Cell Wall Xylans and Arabinoxylans. J. Histochem. Cytochem. 2005, 53, 543–546. [Google Scholar] [CrossRef]
- Zhang, J.; Siika-aho, M.; Tenkanen, M.; Viikari, L. The Role of Acetyl Xylan Esterase in the Solubilization of Xylan and Enzymatic Hydrolysis of Wheat Straw and Giant Reed. Biotechnol. Biofuels 2011, 4, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogorelko, G.; Lionetti, V.; Fursova, O.; Sundaram, R.M.; Qi, M.; Whitham, S.A.; Bogdanove, A.J.; Bellincampi, D.; Zabotina, O.A. Arabidopsis and Brachypodium Distachyon Transgenic Plants Expressing Aspergillus nidulans Acetylesterases Have Decreased Degree of Polysaccharide Acetylation and Increased Resistance to Pathogens. Plant Physiol. 2013, 162, 9–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; He, C.; Zhang, D.; Liu, X.; Xu, Z.; Tian, Y.; Liu, X.-H.; Zang, S.; Pauly, M.; Zhou, Y.; et al. Two Trichome Birefringence-Like Proteins Mediate Xylan Acetylation, Which Is Essential for Leaf Blight Resistance in Rice. Plant Physiol. 2017, 173, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qaseem, M.F.; Wu, A.-M. Balanced Xylan Acetylation Is the Key Regulator of Plant Growth and Development, and Cell Wall Structure and for Industrial Utilization. Int. J. Mol. Sci. 2020, 21, 7875. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.E.; Fry, S.C. Restructuring of Wall-Bound Xyloglucan by Transglycosylation in Living Plant Cells. Plant J. 2001, 26, 23–34. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant Expansins: Diversity and Interactions with Plant Cell Walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989; p. 1546. [Google Scholar]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome Sequence and Analysis of the Tuber Crop Potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A Scaling Normalization Method for Differential Expression Analysis of RNA-Seq Data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeira, F.; Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.; Potter, S.; Finn, R.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucl. Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du Bois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Ralet, M.-C.; Tranquet, O.; Poulain, D.; Moïse, A.; Guillon, F. Monoclonal Antibodies to Rhamnogalacturonan I Backbone. Planta 2010, 231, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An Extended Set of Monoclonal Antibodies to Pectic Homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.E.; Verhertbruggen, Y.; Hervé, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic Homogalacturonan Masks Abundant Sets of Xyloglucan Epitopes in Plant Cell Walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorshkov, V.; Tsers, I.; Islamov, B.; Ageeva, M.; Gogoleva, N.; Mikshina, P.; Parfirova, O.; Gogoleva, O.; Petrova, O.; Gorshkova, T.; et al. The Modification of Plant Cell Wall Polysaccharides in Potato Plants during Pectobacterium atrosepticum-Caused Infection. Plants 2021, 10, 1407. https://doi.org/10.3390/plants10071407
Gorshkov V, Tsers I, Islamov B, Ageeva M, Gogoleva N, Mikshina P, Parfirova O, Gogoleva O, Petrova O, Gorshkova T, et al. The Modification of Plant Cell Wall Polysaccharides in Potato Plants during Pectobacterium atrosepticum-Caused Infection. Plants. 2021; 10(7):1407. https://doi.org/10.3390/plants10071407
Chicago/Turabian StyleGorshkov, Vladimir, Ivan Tsers, Bakhtiyar Islamov, Marina Ageeva, Natalia Gogoleva, Polina Mikshina, Olga Parfirova, Olga Gogoleva, Olga Petrova, Tatyana Gorshkova, and et al. 2021. "The Modification of Plant Cell Wall Polysaccharides in Potato Plants during Pectobacterium atrosepticum-Caused Infection" Plants 10, no. 7: 1407. https://doi.org/10.3390/plants10071407