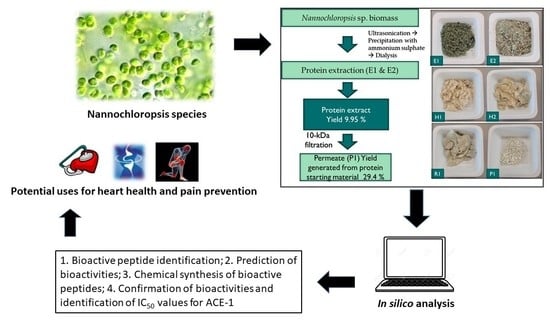
Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Extraction from Supplied Nannochloropsis sp. Biomass
2.3. Xylanase Treatment and Preparation of Molecular Weight Cut Off Permeate Fractions
2.4. Screening and Identification of ACE-1 Inhibitory Activity
2.5. Preparation, Mass Spectrometry Analysis and Identification of Peptides
2.6. In Silico Analysis
2.6.1. Simulated Gastrointestinal (GI) Digestion Using Peptide Cutter
2.6.2. Prediction of Peptide and Peptide Fragment Bioactivities
2.7. Chemical Synthesis, Primary Structure Determination and Theoretical Value Calculation of Peptide Sequences
2.8. Cyclooxygenase Inhibition
2.9. Antimicrobial Screening
3. Results
3.1. Extraction Yields for Protein and Xylanase Treated Permeate Fractions
3.2. Proximate Composition of Whole Alga and Protein Extracts
3.3. ACE-1 Inhibition and Characterisation of Bioactive Peptides Using Mass Spectrometry (MS)
3.4. In Silico Assessment of Identified Peptide Novelity and Additional Bioactivities
3.5. Chemical Synthesis and Confirmation of Bioactivities In Vitro
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darzins, A.; Pienkos, P.; Edye, L. Current Status and Potential for Algal Biofuels Production. In IEA Bioenergy Task 39-CommerCializing Liquid Biofuels; IEA: Paris, France, 2010. [Google Scholar]
- Li, Y.; Horsman, M.; Wu, N.; Lan, C.; Dubois-Calero, N. Biofuels from Microalgae. Biotechnol. Prog. 2008, 24, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Shin, J.H.; Kim, J.D.; Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Cecchin, M.; Berteotti, S.; Paltrinieri, S.; Vigliante, I.; Iadarola, B.; Giovannone, B.; Maffei, M.; Delledonne, M.; Ballottari, M. Improved lipid productivity in Nannochloropsis gaditana in nitrogen-replete conditions by selection of pale green mutants. Biotechnol. Biofuels 2020, 13, 78. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R. The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 1991, 145, 79–99. [Google Scholar] [CrossRef]
- Verspreet, J.; Soetemans, L.; Gargan, C.; Hayes, M.; Bastiaens, L. Nutritional Profiling and Preliminary Bioactivity Screening of Five Micro-Algae Strains Cultivated in Northwest Europe. Foods 2021, 10, 1516. [Google Scholar] [CrossRef]
- Zanella, L.; Vianello, F. Microalgae of the genus Nannochloropsis: Chemical composition and functional implications for human nutrition. J. Funct. Foods 2020, 68, 103919. [Google Scholar] [CrossRef]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.; Romano, G.; Ianora, A. Bioactivity screening of microalgae for antioxidant, anti-inflammatory, anticancer, anti-diabetes and antibacterial activities. Front. Mar. Sci. 2016, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhong-Ji, Q.; Heo, S.-J.; Oh, C.; Kang, D.-H.; Hwa, J.; Park, W.; Choi, I.-W.; Jeon, Y.-J.; Jung, W.-K. Angiotensin I-Converting Enzyme (ACE) Inhibitory Peptide Isolated from Biodiesel Byproducts of Marine Microalgae, Nannochloropsis Oculata. J. Biobased Mater. Bioenergy 2013, 7, 135–142. [Google Scholar] [CrossRef]
- Nacer, W.; Ahmed, F.Z.B.; Merzouk, H.; Benyagoub, O.; Bouanane, S. Evaluation of the anti-inflammatory and antioxidant effects of the microalgae Nannochloropsis gaditana in streptozotocin-induced diabetic rats. J. Diabetes Metab. Disord. 2020, 19, 1483–1490. [Google Scholar] [CrossRef]
- Nasirian, F.; Sarir, H.; Moradi-Kor, N. Antihyperglycemic and antihyperlipidemic activities of Nannochloropsis oculata microalgae in Streptozotocin-induced diabetic rats. Biomol. Concepts 2019, 10, 37–43. [Google Scholar] [CrossRef]
- Werman, M.J.; Sukenik, A.; Mokady, S. Effects of the Marine Unicellular Alga Nannochloropsis sp. to Reduce the Plasma and Liver Cholesterol Levels in Male Rats Fed on Diets with Cholesterol. Biosci. Biotechnol. Biochem. 2014, 67, 2266–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimamoto, K.; Matsuki, T.; Iimura, O. Angiotensin-converting enzyme inhibitors and the kallikrein-kinin system. J. Cardiovasc. Pharmacol. 1990, 15, 83–90. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Peng, C.; Jiao, R.; Wong, Y.M.; Yang, N.; Huang, Y. Anti-hypertensive Nutraceuticals and Functional Foods. J. Agric. Food Chem. 2009, 57, 4485–4499. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Vasbinder, A.; Anderson, E.; Catalan, T.; Shadid, H.R.; Berlin, H.; Padalia, K.; O’Hayer, P.; Meloche, C.; Azam, T.U.; et al. Angiotensin-Converting Enzyme Inhibitors, Angiotensin II Receptor Blockers, and Outcomes in Patients Hospitalized for COVID-19. J. Am. Heart Assoc. 2021, 10, e023535. [Google Scholar] [CrossRef]
- Mohanty, D.; Mohapatra, S.; Misra, S.; Sahu, P. Milk derived bioactive peptides and their impact on human health—A review. Saudi J. Biol. Sci. 2016, 23, 577–583. [Google Scholar] [CrossRef] [Green Version]
- Gallego, M.; Mora, L.; Hayes, M.; Reig, M.; Toldrá, F. Effect of cooking and in vitro digestion on the antioxidant activity of dry-cured ham by-products. Food Res. Int. 2017, 97, 296–306. [Google Scholar] [CrossRef]
- Mora, L.; Hayes, M. Cardioprotective Cryptides Derived from Fish and Other Food Sources: Generation, Application, and Future Markets. J. Agric. Food Chem. 2015, 63, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M. Bioactive Peptides in Preventative Healthcare: An Overview of Bioactivities and Suggested Methods to Assess Potential Applications. Curr. Pharm. Des. 2021, 27, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Abeer, M.M.; Trajkovic, S.; Brayden, D.J. Measuring the oral bioavailability of protein hydrolysates derived from food sources: A critical review of current bioassays. Biomed. Pharmacother. 2021, 144, 112275. [Google Scholar] [CrossRef]
- Jin, J.; Ma, H.; Zhou, C.; Luo, M.; Liu, W.; Qu, W.; He, R.; Luo, L.; Yagoub, A.E.-G.A. Effect of degree of hydrolysis on the bioavailability of corn gluten meal hydrolysates. J. Sci. Food Agric. 2014, 95, 2501–2509. [Google Scholar] [CrossRef]
- Galland-Irmouli, A.; Fleurence, J.; Lamghari, R.; Lucon, M.; Rouxel, C.; Barbaroux, O.; Bronowicki, J.; Villaume, C.; Gueant, J. Nutritional value of proteins from edible seaweed Palmaria palmata (Dulse). J. Nutr. Biochem. 1990, 10, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and characterisation of bioactive pro-peptides with in vitro renin inhibitory activities from the macroalga Palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef] [PubMed]
- Tekkol, G.E.; Sargin, S.; Karaçanci, S.; Pembeci, C.; Mandaci, S.; Akgun, I.H.; Altinel, B.; Ongen, G. Production of GH11 xylanase for bakery industry by solid state fermentation. J. Biotechnol. 2017, 256, S55. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Gritsenko, M.A.; Xu, Z.; Liu, T.; Smith, R.D. Large-Scale and Deep Quantitative Proteome Profiling Using Isobaric Labeling Coupled with Two-Dimensional LC–MS/MS. Methods Mol. Biol. 2016, 1410, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, C.; Aluko, R.E.; Hassain, M.; Rai, D.K.; Hayes, M. Potential of a renin inhibitory peptide from the red seaweed Palmaria palmata as a functional food ingredient following confirmation and characterisation of a hypotensive effect in spontaneously hypertensive rats. J. Agric. Food Chem. 2014, 62, 8352–8356. [Google Scholar] [CrossRef]
- Willenberg, I.; Meschede, A.K.; Gueler, F.; Jang, M.-S.; Shushakova, N.; Schebb, N.H. Food Polyphenols Fail to Cause a Biologically Relevant Reduction of COX-2 Activity. PLoS ONE 2015, 10, e0139147. [Google Scholar] [CrossRef]
- Hayes, M.; Ross, R.P.; Fitzgerald, G.F.; Hill, C.; Stanton, C. Casein-Derived Antimicrobial Peptides Generated by Lactobacillus acidophilus DPC6026. Appl. Environ. Microbiol. 2006, 72, 2260–2264. [Google Scholar] [CrossRef] [Green Version]
- Fleurence, J.; Le Coeur, C.; Mabeau, S.; Maurice, M.; Landrein, A. Comparison of different extractive procedures for proteins from the edible seaweeds Ulva rigida and Ulva rotundata. J. Appl. Phycol. 1995, 7, 577–582. [Google Scholar] [CrossRef]
- Wijers, T.; Hylkema, A.; Visser, T.; Timmermans, K. Effects of preservation on protein extraction in four seaweed species. J. Appl. Phycol. 2020, 32, 3401–3409. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists. Official Methods of Analysis of AOAC International; AOAC International: Rockville, MD, USA, 1998. [Google Scholar]
- Qian, Z.-J.; Heo, S.-J.; Oh, C.H.; Kang, D.-H.; Jeong, S.H.; Park, W.S.; II-Whan, C.; Jeon, Y.-J.; Won, K. Angiotensin 1 converting enzyme inhibitory peptides isolated from Biodiesel By Product of Marine Microalga Nannochloropsis oculata. J. Biobased Mater. BioEnergy 2013, 7, 135–142. [Google Scholar] [CrossRef]
- Samarakoon, K.W.; O-Nam, K.; Ko, J.-Y.; Lee, J.-H.; Kang, M.-C.; Kim, D.; Lee, J.B.; Lee, J.-S.; Jeon, Y.-J. Purification and identification of novel angiotensin-I converting enzyme (ACE) inhibitory peptides from cultured marine microalgae (Nannochloropsis oculata) protein hydrolysate. J. Appl. Phycol. 2013, 25, 1595–1606. [Google Scholar] [CrossRef]
- Sanjeewa, K.K.A.; Fernando, I.P.S.; Samarakoon, K.W.; Lakmal, H.H.C.; Kim, E.-A.; Kwon, O.N.; Dilshara, M.G.; Lee, J.-B.; Jeon, Y.-J. Antiinflammatory and anticancer activities of sterol rich fractions of cultured marine microalga Nannochloropsis oculata. Algae 2016, 31, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Fithriani, D.; Ambarwaty, D.; Nurhayati, D. Identification of bioactive compounds from Nannochloropsis sp. IOP Conf. Ser. Earth Environ. Sci. 2020, 404, 012064. [Google Scholar] [CrossRef]
- Purcell, D.; Packer, M.A.; Hayes, M. Angiotensin-I-Converting Enzyme Inhibitory Activity of Protein Hydrolysates Generated from the Macroalga Laminaria digitata (Hudson) JV Lamouroux 1813. Foods 2022, 11, 1792. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [Green Version]
- Veitch, C.R.; Power, A.S.; Erickson, J.R. CaMKII Inhibition is a Novel Therapeutic Strategy to Prevent Diabetic Cardiomyopathy. Front. Pharmacol. 2021, 12, 1922. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.-R.; Chi, C.-F.; Zhang, Q.-H.; Luo, H.-Y. Preparation and evaluation of antioxidant peptides from ethanol-soluble proteins hydrolysate of Sphyrna lewini muscle. Peptides 2012, 36, 240–250. [Google Scholar] [CrossRef]
- Mojica, L.; Chen, K.; de Mejía, E.G. Impact of Commercial Precooking of Common Bean (Phaseolus vulgaris) on the Generation of Peptides, After Pepsin-Pancreatin Hydrolysis, Capable to Inhibit Dipeptidyl Peptidase-IV. J. Food Sci. 2014, 80, H188–H198. [Google Scholar] [CrossRef]
- Koivunen, E.; Wang, B.; Ruoslahti, E. Phage Libraries Displaying Cyclic Peptides with Different Ring Sizes: Ligand Specificities of the RGD-Directed Integrins. Nat. Biotechnol. 1995, 13, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Ishida, A.; Shigeri, Y.; Tatsu, Y.; Uegaki, K.; Kameshita, I.; Okuno, S.; Kitami, T.; Yumoto, N.; Fujisawa, H. Critical amino acid residues of AIP, a highly specific inhibitory peptide of calmodulin-dependent protein kinase II. FEBS Lett. 1998, 427, 115–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrysson, H.; Hayes, M.; Eimer, F.; Carlsson, N.-G.; Toth, G.B.; Undeland, I. Production of protein extracts from Swedish red, green, and brown seaweeds, Porphyra umbilicalis Kützing, Ulva lactuca Linnaeus, and Saccharina latissima (Linnaeus) J. V. Lamouroux using three different methods. J. Appl. Phycol. 2018, 30, 3565–3580. [Google Scholar] [CrossRef] [Green Version]
- Echave, J.; Fraga-Corral, M.; Garcia-Perez, P.; Popović-Djordjević, J.; Avdović, E.H.; Radulović, M.; Xiao, J.; A. Prieto, M.; Simal-Gandara, J. Seaweed Protein Hydrolysates and Bioactive Peptides: Extraction, Purification, and Applications. Mar. Drugs 2021, 19, 500. [Google Scholar] [CrossRef] [PubMed]
- Mæhre, H.K. Seaweed Proteins—How to Get to Them? Effects of Processing on Nutritional Value, Bioaccessibility and Extractability. Ph.D. Thesis, The Artic University of Norway (UIT), Tromsø, Norway, 2016. Available online: https://hdl.handle.net/10037/9130 (accessed on 30 September 2022).
- Angell, A.R.; Paul, N.; de Nys, R. A comparison of protocols for isolating and concentrating protein from the green seaweed Ulva ohnoi. J. Appl. Phycol. 2017, 29, 1011–1026. [Google Scholar] [CrossRef]
- DE Amorim, A.P.; DA Silva, G.H.; Brandão, R.M.P.; Porto, A.L.F.; Bezerra, R.P. Algae as a source of peptides inhibitors of the angiotensin-converting enzyme: A systematic review. Ann. Braz. Acad. Sci. 2022, 94, e20201636. [Google Scholar] [CrossRef]
- Goddard-Borger, E.D.; Sakaguchi, K.; Reitinger, S.; Watanabe, N.; Ho, M.; Withers, S.G. Mechanistic insights into the 1,3 xylanases: Useful enzymes for manipulation of algal biomass. J. Am. Chem. Soc. 2012, 134, 3895–3902. [Google Scholar] [CrossRef]
- Kroger, M.; Klemm, M.; Nelles, M. Hydrothermal disintegration and extraction of different microalgal species. Energies 2018, 11, 450. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Arantes, V.; Saddler, J.N. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: Is it an additive or synergistic effect? Biotechnol. Biofuels 2011, 4, 36. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, S.; Singh, B. Microbial Xylanases in Bread Making. In Reference Module in Food Science; Academic Press: Oxford, UK, 2019; pp. 140–149. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Oh, D.H.; Lee, B.H. Bioactive peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef]
- Daskaya-Dikmen, C.; Yucetepe, A.; Karbancioglu-Guler, F.; Daskaya, H.; Ozcelik, B. Angiotensin-I-Converting Enzyme (ACE)-Inhibitory Peptides from Plants. Nutrients 2017, 9, 316. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Y.; Zhang, J.-T.; Miyakawa, T.; Li, G.-M.; Gu, R.-Z.; Tanokura, M. Antioxidant properties and inhibition of angiotensin-converting enzyme by highly active peptides from wheat gluten. Sci. Rep. 2021, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.A.; Coscueta, E.R.; Nova, P.; Silva, J.L.; Pintado, M.M. Bioactive Hydrolysates from Chlorella vulgaris: Optimal Process and Bioactive Properties. Molecules 2022, 27, 2505. [Google Scholar] [CrossRef]
- Epellicena, P.; Eschulman, H. CaMKII inhibitors: From research tools to therapeutic agents. Front. Pharmacol. 2014, 5, 21. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Roa, A.; Espinoza-Culupú, A.; Torres-García, O.; Borges, M.M.; Avino, I.N.; Alves, F.L.; Miranda, A.; Patarroyo, M.A.; da Silva, P.I.; Bello, F. Sarconesin II, a New Antimicrobial Peptide Isolated from Sarconesiopsis magellanica Excretions and Secretions. Molecules 2019, 24, 2077. [Google Scholar] [CrossRef] [PubMed]




| Species | Proteins [% DW] | Lipids [% DW] | Carbohydrates [% DW] | Other [% DW] | References |
|---|---|---|---|---|---|
| Nannochloropsis sp. biomass (this study) Nannochloropsis sp. protein extract (this study) | 27.95 ± 1.42 26.16 + 0.4 | 1.25 ± 0.39 0.38 ± 0.3 | n/d n/d | Ash 23.18 ± 2.59; Moisture 6.37 ± 0.75 27.78 ± 0.13 | This study This study |
| Nannochloropsis gaditana | 46.2 ± 0.3 | 31.9 ± 0.6 | 15.8 ± 0.4 | Ash 6.4 ± 0.0 | Verspreet et al., 2021 [6] |
| Nannochloropsis oculata | 30.4 ± 1.8 | 20.5 ± 1.2 | 37.1 ± 1.7 | Mineral, fiber, etc. 11.1 ± 0.8 | Qian et al., 2013 [34] |
| N. oculata | 31.0 ± 0.0 | 1.3 ± 0.0 | 17.8 ± 0.1 | Ash 32.9 ± 0.0 Fiber 4.4 ± 0.0 | Samarakoon et al., 2013 [35] |
| N. oculata | 30.5 | 8.0 | 19.6 | Ash 30.6 | Sanjeewa et al., 2016 [36] |
| N. gaditana | 28.0 | 18.4 | 45.0 | - | Nacer et al., 2020 [10] |
| Nannochloropsis sp. | 31.7 ± 0.1 | 15.0 ± 0.1 | 9.0 ± 0.4 | Ash 27.3 ± 0.2 | Fithriani et al., 2020 [37] |
| Parent Protein Name & UniProt Accession Number | Peptide Single Amino Acid Sequence | Peptide Ranker Value 1 | Novelty (Found in Database 1,2) | Observed Bioactivity In Vitro | Simulated Digestion Using PeptideCutter 3/Peptide Digestion Fragments | Associated Predicted Bioactivities of Peptide Fragments Resulting from Simulated GI Digestion | References |
|---|---|---|---|---|---|---|---|
| Chlorophyll a-b binding protein, chloroplastic OS = Porphyridium purpureum OX = 35,688 GN = FVE85_6435 PE = 4 SV = 1; Chlorophyll a-b binding protein 1B-21, chloroplastic OS = Porphyridium purpureum OX = 35,688 GN = FVE85_3955 PE = 4 SV = 1 | AGDVGFDPLGF | 0.89 | Novel | ACE-1 inhibitory, Antibacterial activity against E. coli at a concentration of 1 mg/mL | Chymotrypsin–position 6 and 9. Peptide fragments AGDVGF, DPL and GF | No predicted bioactivities associated with any peptide fragments reported in BIOPEP | [26] |
| Mannose-6-phosphate isomerase OS = Vibrio sp. SM1977 OX = 2,662,262 GN = manA PE = 3 SV = 1; Uncharacterized protein OS = Porphyridium purpureum OX = 35,688 GN = FVE85_9013 PE = 4 SV = 1; REVERSED p-aminobenzoic acid synthase OS = Ectocarpus siliculosus OX = 2880 GN = PABS PE = 4 SV = 1; REVERSED Uncharacterized protein OS = Gimesia algae OX = 2,527,971 GN = Pan161_23400 PE = 4 SV = 1; REVERSED NADH-ubiquinone oxidoreductase chain 4 OS = Guillardia theta OX = 55,529 GN = nad4 PE = 3 SV = 1; REVERSED NADH-ubiquinone oxidoreductase chain 4 OS = Chroomonas placoidea OX = 173,977 GN = nad4 PE = 3 SV = 1; REVERSED Transcriptional regulator OS = Formosa algae OX = 225,843 GN = BKP44_19185 PE = 4 SV = 1; REVERSED Deoxyguanosine kinase OS = Shewanella algae OX = 38,313 GN = BFS86_13800 PE = 4 SV = 1 | GDVGLF | 0.82 | Novel | ACE-1 | Pepsin position 4, 5, 6, Chymotrypsin position 5 and 6. Peptide fragments GDVG, L, F | No predicted bioactivities associated with peptide fragments reported in BIOPEP | [26] |
| Uncharacterized protein OS = Mesonia algae OX = 213,248 GN = LX95_02145 PE = 4 SV = 1 | YANDLLCMPI | 0.77 | Novel | ACE-1; COX-1 and COX-2 inhibition | Pepsin 1, 4, 5, 6 and Chymotrypsin at position 4, 5 and 6. Peptide fragments Y, AND, L, L, CMPI | MPI corresponds to amino acid fragments 2–5 of the antithrombotic peptide DMPIQAFLLYQEPVLGPVR, AND has sequence similarity with peptides NDQF an antiviral peptide and NDPQF a bitterness suppressing peptide but no reported exact matches for AND | [26] |
| REVERSED SHR-BD domain-containing protein OS = Ectocarpus siliculosus OX = 2880 GN = Esi_0004_0132 PE = 4 SV = 1 | KGGGSGANGGRL | 0.72 | Novel | ACE-1 | Pepsin, Trypsin and Chymotrypsin cut the peptide at positions 1, 11 and 12. Resulting peptide fragments are K, GGGSGANGGR, L | GGGSGANGGR has sequence similarity in terms of the amino acids GR with peptides GGAAGGR a DPP-IV inhibitory peptide, TKHGGRINTL an antiviral peptide and the ACE inhibitory peptide FPVGRGL | [26] |
| Phycocyanin protein, accession number: tr|S5FXR4|S5FXR4_9CYAN | NKFPYTTQ | 0.19 | Novel | ACE-1 inhibition | Trypsin–position 2 Pepsin–position 3 Peptide fragment: NK, F, PYTTQ | NK–ACE-1 inhibitor found previously in Wakame seaweed; TTQ found as an ACE inhibitor previously isolated from bean (Phaseolus vulgaris) | [41,42] |
| Protein R-phycocyanin alpha subunit Synechococcus sp. (accession number: P27288 PHCA_SYNPW | VYNKFPYTTQ | 0.18 | Novel | ACE-1 inhibition | Trypsin–position 4 Pepsin–1, 2, 5 Resulting peptide fragments: V, Y, NK, F, PYTTQ | NK–ACE inhibitor found previously in Wakame seaweed; TTQ found as an ACE inhibitor previously isolated from bean (Phaseolus vulgaris) | [43] |
| Transcriptor initiation protein from Porphyridium sp. (accession number tr|A0A5J4YUD5|A0A5J4YUD5_PORPP | LVGADAHALGVICS | 0.39 | Novel | ACE-1 inhibition | Pepsin–positions 1 and 8 Resulting peptide fragments: L, VGADAHA, LGIVICS | AHA, also occurs at the C-terminal end of a peptide NGRAHA that has antithrombotic activity; | [44] |
| HEAT repeat-containing protein 1 OS = Ectocarpus siliculosus OX = 2880 GN = Esi_0125_0072 PE = 3 SV = 1 (accession number: tr|D7FJ23|D7FJ23_ECTSI) | VVGAVGAADLL | 0.23 | Novel | ACE-1 inhibition | Pepsin at positions 9, 10 and 11 Fragments: VVGAVGAAD, L, L | AAD found at C-terminal end of CAMKII inhibitor KKALRRQEAADAL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, M.; Mora, L.; Lucakova, S. Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis. Biomolecules 2022, 12, 1806. https://doi.org/10.3390/biom12121806
Hayes M, Mora L, Lucakova S. Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis. Biomolecules. 2022; 12(12):1806. https://doi.org/10.3390/biom12121806
Chicago/Turabian StyleHayes, Maria, Leticia Mora, and Simona Lucakova. 2022. "Identification of Bioactive Peptides from Nannochloropsis oculata Using a Combination of Enzymatic Treatment, in Silico Analysis and Chemical Synthesis" Biomolecules 12, no. 12: 1806. https://doi.org/10.3390/biom12121806