Dilignans with a Chromanol Motif Discovered by Molecular Networking from the Stem Barks of Magnolia obovata and Their Proprotein Convertase Subtilisin/Kexin Type 9 Expression Inhibitory Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
2.2. Plant Material
2.3. MS/MS Molecular Networking
2.4. Extraction and Isolation
2.5. Cell Culture, Drugs and Chemicals
2.6. Quantitative Real-Time RT-PCR
2.7. Immunoblot Analysis
2.8. Statistical Analysis
2.9. Selection of Candidate Genes and Construction of Protein-Protein Interaction Network
2.10. ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) Analysis
3. Results and Discussion
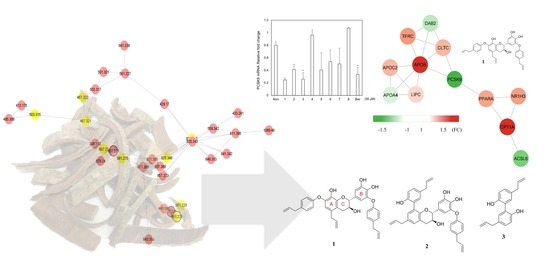
3.1. Dereplication Using GNPS Molecular Networking and Network Annotation Propagation (NAP)
3.2. Isolation and Structural Characterization
3.3. Biological Assay for PCSK9 mRNA Inhibitory Effect
3.4. Construction of Protein-Protein Interaction Network of Candidate Genes
3.5. ADMET Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| MN | Molecular networking |
| LDL-C | Low-density lipoprotein cholesterol |
| PCSK9 | Proprotein convertase subtilisin/kexin type 9 |
| LDLR | Low-density lipoprotein receptor |
| GNPS | Global Natural Products Social Molecular Networking |
| NAP | Network Annotation Propagation |
| HRESIMS | High resolution-electrospray ionization-mass spectroscopy |
| SEM | Standard error of the mean |
| STRING | Search Tool for the Retrieval of Interacting Genes |
| ADMET | Absorption, Distribution, Metabolism, Excretion and Toxicity |
| MFs | Molecular families |
| VDss | Steady state Volume of Distribution |
| CYP | Cytochrome P 450 enzyme |
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Gaudencio, S.P.; Pereira, F. Dereplication: Racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779. [Google Scholar] [CrossRef]
- Kang, K.B.; Woo, S.; Ernst, M.; van der Hooft, J.J.J.; Nothias, L.-F.; da Silva, R.R.; Dorrestein, P.C.; Sung, S.H.; Lee, M. Assessing specialized metabolite diversity of Alnus species by a digitized LC—MS/MS data analysis workflow. Phytochemistry 2020, 173, 112292. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Kang, K.B.; Kim, J.; Sung, S.H. Molecular networking reveals the chemical diversity of selaginellin derivatives, natural phosphodiesterase-4 inhibitors from Selaginella tamariscina. J. Nat. Prod. 2019, 82, 1820–1830. [Google Scholar] [CrossRef] [PubMed]
- Marner, M.; Patras, M.A.; Kurz, M.; Zubeil, F.; Förster, F.; Schuler, S.; Bauer, A.; Hammann, P.; Vilcinskas, A.; Schäberle, T.F.; et al. Molecular networking-guided discovery and characterization of stechlisins, a group of cyclic lipopeptides from a Pseudomonas sp. J. Nat. Prod. 2020, 83, 2607–2617. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Allard, P.-M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.-F.; Cachet, X.; et al. Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Nothias, L.-F.; Nothias-Esposito, M.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nothias-Esposito, M.; Nothias, L.F.; da Silva, R.R.; Retailleau, P.; Zhang, Z.; Leyssen, P.; Roussi, F.; Touboul, D.; Paolini, J.; Dorrestein, P.C.; et al. Investigation of premyrsinane and myrsinane esters in Euphorbia cupanii and Euphobia pithyusa with MS2LDA and combinatorial molecular network annotation propagation. J. Nat. Prod. 2019, 82, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS ONE 2018, 14, e1006089. [Google Scholar] [CrossRef]
- Yi, Z.; Zhu, Z.-J. Overview of tandem mass spectral and metabolite databases for metabolite identification in metabolomics. In Computational Methods and Data Analysis for Metabolomics; Li, S., Ed.; Springer: New York, NY, USA, 2014; pp. 139–148. ISBN 9781071602386. [Google Scholar]
- Alla, V.M.; Agrawal, V.; DeNazareth, A.; Mohiuddin, S.; Ravilla, S.; Rendell, M. A reappraisal of the risks and benefits of treating to target with cholesterol lowering drugs. Drugs 2013, 73, 1025–1054. [Google Scholar] [CrossRef] [PubMed]
- Rosenson, R.S.; Hegele, R.A.; Fazio, S.; Cannon, C.P. The evolving future of PCSK9 inhibitors. J. Am. Coll. Cardiol. 2018, 72, 314–329. [Google Scholar] [CrossRef] [PubMed]
- Kotowski, I.K.; Pertsemlidis, A.; Luke, A.; Cooper, R.S.; Vega, G.L.; Cohen, J.C.; Hobbs, H.H. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 2006, 78, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.; Kim, Y.; Chae, H.; Choi, Y.H.; Ahn, H.; Yoo, H.; Kang, M.; Kim, J.; Chin, Y.-W. Prenylated flavonoids from the roots and rhizomes of Sophora tonkinensis and their effects on the expression of inflammatory mediators and proprotein convertase subtilisin/kexin type 9. J. Nat. Prod. 2019, 82, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Hang, P.; Hu, J.; Zheng, Y.; Sun, H.; Guo, J.; Liu, K.; Du, Z. Aloe-emodin exerts cholesterol-lowering effects by inhibiting proprotein convertase subtilisin/kexin type 9 in hyperlipidemic rats. Acta Pharmacol. Sin. 2020, 41, 1085–1092. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Zhang, X.; Li, J.; Xi, C.; Wang, W.; Lu, Y.; Xuan, L. 23,24-Dihydrocucurbitacin B promotes lipid clearance by dual transcriptional regulation of LDLR and PCSK9. Acta Pharmacol. Sin. 2020, 41, 327–335. [Google Scholar] [CrossRef]
- Dong, B.; Li, H.; Singh, A.B.; Cao, A.; Liu, J. Inhibition of PCSK9 transcription by berberine involves down-regulation of hepatic HNF1α protein expression through the ubiquitin-proteasome degradation pathway. J. Biol. Chem. 2015, 290, 4047–4058. [Google Scholar] [CrossRef] [Green Version]
- Pel, P.; Chae, H.-S.; Nhoek, P.; Kim, Y.-M.; Khiev, P.; Kim, G.J.; Nam, J.-W.; Choi, H.; Choi, Y.H.; Chin, Y.-W. A stilbene dimer and flavonoids from the aerial parts of Chromolaena odorata with proprotein convertase subtilisin/kexin type 9 expression inhibitory activity. Bioorg. Chem. 2020, 99, 103869. [Google Scholar] [CrossRef]
- Baek, Y.-S.; Seo, K.-H.; Lee, D.-Y.; Kwon, O.-K.; Baek, N.-I. A new neolignan, isoobovatol, from the fruits of Magnolia obovata. Chem. Nat. Compd. 2016, 52, 986–988. [Google Scholar] [CrossRef]
- Seo, K.-H.; Lee, D.-Y.; Lee, Y.-G.; Baek, N.-I. Dineolignans of 3-O-4′ diphenyl ether-type from fruits of Magnolia obovata. Phytochemistry 2017, 136, 133–140. [Google Scholar] [CrossRef]
- Matsuda, H.; Kageura, T.; Oda, M.; Morikawa, T.; Sakamoto, Y.; Yoshikawa, M. Effects of constituents from the bark of Magnolia obovata on nitric oxide production in lipopolysaccharide-activated macrophages. Chem. Pharm. Bull. 2001, 49, 716–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patočka, J.; Jakl, J.; Strunecká, A. Expectations of biologically active compounds of the genus Magnolia in biomedicine. J. Appl. Biomed. 2006, 4, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Sarrica, A.; Kirika, N.; Romeo, M.; Salmona, M.; Diomede, L. Safety and Toxicology of Magnolol and Honokiol. Planta Med. 2018, 84, 1151–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.J.; Moon, J.S.; Kim, S.I.; Bahn, Y.-S.; Lee, H.; Kang, T.H.; Shin, H.M.; Kim, S.U. A phenylpropanoid glycoside as a calcineurin inhibitor isolated from Magnolia obovata Thunb. J. Microbiol. Biotechnol. 2015, 25, 1429–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, K.-H.; Lee, D.-Y.; Jung, J.-W.; Lee, D.-S.; Kim, Y.-C.; Lee, Y.-H.; Baek, N.-I. Neolignans from the Fruits of Magnolia obovata inhibit NO production and have neuroprotective effects. Helv. Chim. Acta 2016, 99, 411–415. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.B.; Park, E.J.; da Silva, R.R.; Kim, H.W.; Dorrestein, P.C.; Sung, S.H. Targeted isolation of neuroprotective dicoumaroyl neolignans and lignans from Sageretia theezans using in silico molecular network annotation propagation-based dereplication. J. Nat. Prod. 2018, 81, 1819–1828. [Google Scholar] [CrossRef]
- Chae, H.-S.; You, B.H.; Kim, D.-Y.; Lee, H.; Ko, H.W.; Ko, H.-J.; Choi, Y.H.; Choi, S.S.; Chin, Y.-W. Sauchinone controls hepatic cholesterol homeostasis by the negative regulation of PCSK9 transcriptional network. Sci. Rep. 2018, 8, 6737. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Lagorce, D.; Douguet, D.; Miteva, M.A.; Villoutreix, B.O. Computational analysis of calculated physicochemical and ADMET properties of protein-protein interaction inhibitors. Sci. Rep. 2017, 7, 46277. [Google Scholar] [CrossRef]
- Lee, S.-K.; Kim, H.-N.; Kang, Y.-R.; Lee, C.W.; Kim, H.-M.; Han, D.C.; Shin, J.; Bae, K.; Kwon, B.-M. Obovatol inhibits colorectal cancer growth by inhibiting tumor cell proliferation and inducing apoptosis. Bioorg. Med. Chem. 2008, 16, 8397–8402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wei, H.; Jiang, Z.; Li, X.; Jiao, K.; Jia, X.; Hou, Y.; Li, N. Natural potential neuroinflammatory inhibitors from Alhagi sparsifolia Shap. Bioorg. Med. Chem. Lett. 2017, 27, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Han, F.-Y.; Lu, L.-W.; Yao, G.-D.; Zhang, Y.-Y.; Wang, X.-B.; Lin, B.; Huang, X.-X.; Song, S.-J. Isolation of enantiomeric furolactones and furofurans from Rubus idaeus L. with neuroprotective activities. Phytochemistry 2019, 164, 122–129. [Google Scholar] [CrossRef]
- Shen, C.-C.; Chang, Y.-S.; Ho, L.-K. Nuclear magnetic resonance studies of 5,7-dihydroxyflavonoids. Phytochemistry 1993, 34, 843–845. [Google Scholar] [CrossRef]
- Rinaldo, D.; Batista, J.M., Jr.; Rodrigues, J.; Benfatti, A.C.; Rodrigues, C.M.; Dos Santos, L.C.; Furlan, M.; Vilegas, W. Determination of catechin diastereomers from the leaves of Byrsonima species using chiral HPLC-PAD-CD. Chirality 2010, 22, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Sebastiano, M.R.; Konstantinidou, G. Targeting long chain acyl-Coa synthetases for cancer therapy. Int. J. Mol. Sci. 2019, 20, 3624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnefont, J.-P.; Djouadi, F.; Prip-Buus, C.; Gobin, S.; Munnich, A.; Bastin, J. Carnitine palmitoyltransferases 1 and 2: Biochemical, molecular and medical aspects. Mol. Asp. Med. 2004, 25, 495–520. [Google Scholar] [CrossRef]
- Drouin-Chartier, J.-P.; Tremblay, A.J.; Hogue, J.-C.; Lemelin, V.; Lamarche, B.; Couture, P. Plasma PCSK9 correlates with apoB-48-containing triglyceride-rich lipoprotein production in men with insulin resistance. J. Lipid Res. 2018, 59, 1501–1509. [Google Scholar] [CrossRef] [Green Version]
- Jeyaraman, P.; Samuel, M.; Johnson, A.; Raman, N. Synthesis, characterization, ADMET, in vitro and in vivo studies of mixed ligand metal complexes from a curcumin Schiff base and lawsone. Nucleosides Nucleotides Nucleic Acids 2020, 40, 242–263. [Google Scholar] [CrossRef]
- Willrich, M.A.V.; Hirata, M.H.; Hirata, R.D.C. Statin regulation of CYP3A4 and CYP3A5 expression. Pharmacogenomics 2009, 10, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Ming, E.E.; Davidson, M.H.; Gandhi, S.K.; Marotti, M.; Miles, C.G.; Ke, X.; McKenney, J.M. Concomitant use of statins and CYP3A4 inhibitors in administrative claims and electronic medical records databases. J. Clin. Lipidol. 2008, 2, 453–463. [Google Scholar] [CrossRef] [PubMed]







| Obovatolin A (1) | Obovatolin B (2) | |||||
|---|---|---|---|---|---|---|
| Position | δC | Type | δH (J in Hz) | δC | Type | δH (J in Hz) |
| 2 | 82.1 | CH | 4.80 d (6.3) | 82.1 | CH | 4.79 d (5.6) |
| 3 | 68.4 | CH | 4.10 m | 68.4 | CH | 4.07 m |
| 4a | 30.3 | CH2 | 2.83 dd (16.4, 5.0) | 32.8 | CH2 | 2.91 dd (16.4, 4.7) |
| 4b | 2.66 dd (16.4, 6.8) | 2.77 dd (16.4, 6.3) | ||||
| 5 | 130.2 | C | 130.3 | CH | 6.88 d (2.2) | |
| 6 | 115.1 | CH | 6.34 s | 133.1 | C | |
| 7 | 143.0 | C | 131.1 | CH | 6.84 d (2.2) | |
| 8 | 136.9 | C | 127.7 | C | ||
| 9 | 144.6 | C | 150.6 | C | ||
| 10 | 116.8 | C | 121.5 | C | ||
| 1′ | 131.3 | C | 131.6 | C | ||
| 2′ | 111.2 | CH | 6.46 d (1.9) | 110.7 | CH | 6.36 d (2.2) |
| 3′ | 145.6 | C | 145.3 | C | ||
| 4′ | 138.0 | C | 137.6 | C | ||
| 5′ | 148.1 | C | 147.9 | C | ||
| 6′ | 110.4 | CH | 6.71 d (1.9) | 110.0 | CH | 6.61 d (2.2) |
| 1″ | 37.4 | CH2 | 3.19 d (6.2) | 40.49 | CH2 | 3.33 overlap |
| 2″ | 137.8 | CH | 5.84 m | 139.3 | CH | 5.96 overlap |
| 3″ | 116.0 | CH2 | 4.92 m | 115.8 | CH2 | 5.05 m |
| 1‴ | 134.9 | C | 132.1 | C | ||
| 2‴ | 130.5 | CH | 7.06 d (8.6) | 132.5 | CH | 6.70 d (2.0) |
| 3‴ | 117.7 | CH | 6.78 d (8.6) | 127.0 | C | |
| 4‴ | 158.1 | C | 153.7 | C | ||
| 5‴ | 117.7 | CH | 6.78 d (8.6) | 116.8 | CH | 6.72 d (8.2) |
| 6‴ | 130.5 | CH | 7.06 d (8.6) | 129.5 | CH | 6.91 dd (8.2, 2.0) |
| 7‴ | 40.41 | CH2 | 3.32 overlap | 40.4 | CH2 | 3.20 d (6.8) |
| 8‴ | 139.1 | CH | 5.94 overlap | 139.6 | CH | 5.89 m |
| 9‴ | 115.8 | CH2 | 4.98 m | 115.4 | CH2 | 4.98 m |
| 1″″ | 135.5 | C | 135.3 | C | ||
| 2″″ | 130.6 | CH | 7.08 d (8.6) | 130.6 | CH | 7.04 d (8.5) |
| 3″″ | 118.4 | CH | 6.86 d (8.6) | 118.3 | CH | 6.76 d (8.5) |
| 4″″ | 157.5 | C | 157.4 | C | ||
| 5″″ | 118.4 | CH | 6.86 d (8.6) | 118.3 | CH | 6.76 d (8.5) |
| 6″″ | 130.6 | CH | 7.08 d (8.6) | 130.6 | CH | 7.04 d (8.5) |
| 7″″ | 40.39 | CH2 | 3.32 overlap | 40.46 | CH2 | 3.33 overlap |
| 8″″ | 139.2 | CH | 5.93 overlap | 139.1 | CH | 5.95 overlap |
| 9″″ | 115.7 | CH2 | 5.03 m | 115.7 | CH2 | 5.04 m |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, J.; Chae, H.-S.; Pel, P.; Kim, Y.-M.; Choi, Y.H.; Kim, J.; Chin, Y.-W. Dilignans with a Chromanol Motif Discovered by Molecular Networking from the Stem Barks of Magnolia obovata and Their Proprotein Convertase Subtilisin/Kexin Type 9 Expression Inhibitory Activity. Biomolecules 2021, 11, 463. https://doi.org/10.3390/biom11030463
Ahn J, Chae H-S, Pel P, Kim Y-M, Choi YH, Kim J, Chin Y-W. Dilignans with a Chromanol Motif Discovered by Molecular Networking from the Stem Barks of Magnolia obovata and Their Proprotein Convertase Subtilisin/Kexin Type 9 Expression Inhibitory Activity. Biomolecules. 2021; 11(3):463. https://doi.org/10.3390/biom11030463
Chicago/Turabian StyleAhn, Jongmin, Hee-Sung Chae, Pisey Pel, Young-Mi Kim, Young Hee Choi, Jinwoong Kim, and Young-Won Chin. 2021. "Dilignans with a Chromanol Motif Discovered by Molecular Networking from the Stem Barks of Magnolia obovata and Their Proprotein Convertase Subtilisin/Kexin Type 9 Expression Inhibitory Activity" Biomolecules 11, no. 3: 463. https://doi.org/10.3390/biom11030463