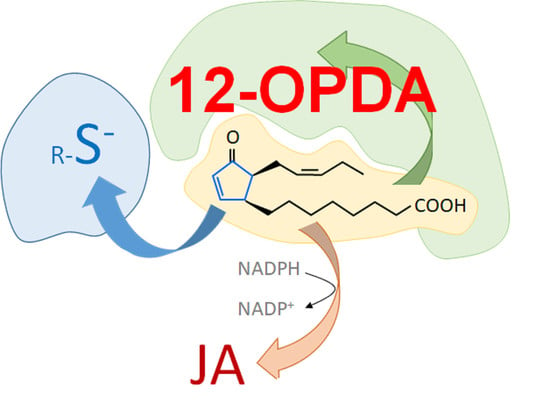
The In Vitro Interaction of 12-Oxophytodienoic Acid and Related Conjugated Carbonyl Compounds with Thiol Antioxidants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Data Processing, Databases and in Silico Studies
2.3. Synthesis and Detection of Cys-12-OPDA
2.4. Mass Spectrometric and Calorimetric Detection of Cysteine Interacting with Abscisic Acid
2.5. Analysis of Free Thiol Content of GSH Incubated with CCCs
2.6. Effect of Exogenously Applied Methylvinyl Ketone (MVK) Incubated with Thiol and Non-thiol Antioxidant on Photochemical Activity of Leaf Discs
2.7. Synthesis and Purification of FBPase, TRX-f1, TRX-m1, 2-CysPRX, NTRC, Cyp20-3 and Cyp20-3 Variants
2.8. Fluorescence Properties of TRX-Fold Proteins and Cyp20-3
2.9. Covalent Modification of TRX-f1 by 12-OPDA for Analysis by Mass Spectrometry
2.10. FBPase Activity Test
2.11. Protein Thiol Estimation with DTNB
2.12. NTRC-Dependent Peroxidase Activity of 2-CysPRX in Presence of CCCs
2.13. ITC-Analysis of Interaction between 12-OPDA and Cyp20-3
2.14. Activity Assays for Cyp20-3 in the Presence of 12-OPDA
3. Results
3.1. Reactivity of Michael Systems with Cysteine, Glutathione and Ascorbate
3.2. The Interaction of 12-OPDA with Plant Thioredoxin
3.3. The Interaction of 12-OPDA with Cyclophilin 20-3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davoine, C.; Falletti, O.; Douki, T.; Iacazio, G.; Ennar, N.; Montillet, J.-L.; Triantaphylidès, C. Adducts of Oxylipin Electrophiles to Glutathione Reflect a 13 Specificity of the Downstream Lipoxygenase Pathway in the Tobacco Hypersensitive Response. Plant Physiol. 2006, 140, 1484–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esterbauer, H.; Zöllner, H.; Scholz, N. Reaction of Glutathione with Conjugated Carbonyls. Z. Nat. C 1975, 30, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The Lipoxygenase Pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef]
- Pierotti, D.; Wofsy, S.C.; Jacob, D.; Rasmussen, R.A. Isoprene and its oxidation products: Methacrolein and methyl vinyl ketone. J. Geophys. Res. Atmos. 1990, 95, 1871–1881. [Google Scholar] [CrossRef]
- Kai, H.; Hirashima, K.; Matsuda, O.; Ikegami, H.; Winkelmann, T.; Nakahara, T.; Iba, K. Thermotolerant cyclamen with reduced acrolein and methyl vinyl ketone. J. Exp. Bot. 2012, 63, 4143–4150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, G.; Brunetti, C.; Tattini, M.; Romano, A.; Biasioli, F.; Tognetti, R.; Loreto, F.; Ferrini, F.; Centritto, M. Dissecting the role of isoprene and stress-related hormones (ABA and ethylene) in Populus nigra exposed to unequal root zone water stress. Tree Physiol. 2017, 37, 1637–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-W.; Li, W.; Viehhauser, A.; He, B.; Kim, S.; Nilsson, A.K.; Andersson, M.X.; Kittle, J.D.; Ambavaram, M.M.R.; Luan, S.; et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9559–9564. [Google Scholar] [CrossRef] [Green Version]
- Rajab, H.; Khan, M.S.; Malagoli, M.; Hell, R.; Wirtz, M. Sulfate-Induced Stomata Closure Requires the Canonical ABA Signal Transduction Machinery. Plants 2019, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Maynard, D.; Gröger, H.; Dierks, T.; Dietz, K.-J. The function of the oxylipin 12-oxophytodienoic acid in cell signaling, stress acclimation, and development. J. Exp. Bot. 2018, 69, 5341–5354. [Google Scholar] [CrossRef]
- Mano, J.; Biswas, S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levonen, A.-L.; Landar, A.; Ramachandran, A.; Ceaser, E.K.; Dickinson, D.A.; Zanoni, G.; Morrow, J.D.; Darley-Usmar, V.M. Cellular mechanisms of redox cell signalling: Role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004, 378, 373–382. [Google Scholar] [CrossRef] [Green Version]
- Burstein, S.H. The chemistry, biology and pharmacology of the cyclopentenone prostaglandins. Prostaglandins Other Lipid Mediat. 2020, 148, 106408. [Google Scholar] [CrossRef]
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575. [Google Scholar] [CrossRef] [Green Version]
- Dueckershoff, K.; Mueller, S.; Mueller, M.J.; Reinders, J. Impact of cyclopentenone-oxylipins on the proteome of Arabidopsis thaliana. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2008, 1784, 1975–1985. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Schaur, R.J.; Siems, W.; Bresgen, N.; Eckl, P.M. 4-Hydroxy-nonenal—A Bioactive Lipid Peroxidation Product. Biomolecules 2015, 5, 2247–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, J.; Bansal, R.; Post, E.; De Jager-Krikken, A.; Hooge, M.N.L.-D.; Poelstra, K. Albumin-Binding and Tumor Vasculature Determine the Antitumor Effect of 15-Deoxy-Δ12,14-Prostaglandin-J2in vivo. Neoplasia 2009, 11, 1348–1358. [Google Scholar] [CrossRef] [Green Version]
- Shibata, T.; Yamada, T.; Ishii, T.; Kumazawa, S.; Nakamura, H.; Masutani, H.; Yodoi, J.; Uchida, K. Thioredoxin as a Molecular Target of Cyclopentenone Prostaglandins. J. Biol. Chem. 2003, 278, 26046–26054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maynard, D.; Müller, S.M.; Hahmeier, M.; Löwe, J.; Feussner, I.; Gröger, H.; Viehhauser, A.; Dietz, K.-J. One-pot synthesis of bioactive cyclopentenones from α-linolenic acid and docosahexaenoic acid. Bioorganic Med. Chem. 2018, 26, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 28 January 2021).
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, A Protein-Small Molecule Docking Web Service Based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. Available online: http://www.swissdock.ch/docking (accessed on 28 January 2021). [CrossRef] [Green Version]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Grimm, M.; Dai, W.-T.; Hou, M.-C.; Xiao, Z.-X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2019, 41, 138–144. [Google Scholar] [CrossRef]
- Pubchem. Available online: pubchem.ncbi.nlm.nih.gov (accessed on 4 December 2020).
- Molinspiration. Available online: molinspiration.com (accessed on 4 December 2020).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.’e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Rascher, U.; Liebig, M.; Luttge, U. Evaluation of instant light-response curves of chlorophyll fluorescence parameters obtained with a portable chlorophyll fluorometer on site in the field. Plant Cell Environ. 2000, 23, 1397–1405. [Google Scholar] [CrossRef]
- Vaseghi, M.-J.; Chibani, K.; Telman, W.; Liebthal, M.F.; Gerken, M.; Schnitzer, H.; Mueller, S.M.; Dietz, K.-J. The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. eLife 2018, 7, e38194. [Google Scholar] [CrossRef]
- Dreyer, A.; Schackmann, A.; Kriznik, A.; Chibani, K.; Wesemann, C.; Vogelsang, L.; Beyer, A.; Dietz, K.-J. Thiol Redox Regulation of Plant β-Carbonic Anhydrase. Biomolecules 2020, 10, 1125. [Google Scholar] [CrossRef] [PubMed]
- Laxa, M.; König, J.; Dietz, K.-J.; Kandlbinder, A. Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis–trans isomerase and redox-related functions. Biochem. J. 2006, 401, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Liebthal, M.; Strüve, M.; Li, X.; Hertle, Y.; Maynard, D.; Hellweg, T.; Viehhauser, A.; Dietz, K.-J. Redox-Dependent Conformational Dynamics of Decameric 2-Cysteine Peroxiredoxin and its Interaction with Cyclophilin 20-3. Plant Cell Physiol. 2016, 57, 1415–1425. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, M.; Michelet, L.; Massot, V.; Trost, P.; Lemaire, S.D. Biochemical Characterization of Glutaredoxins from Chlamydomonas reinhardtii Reveals the Unique Properties of a Chloroplastic CGFS-type Glutaredoxin. J. Biol. Chem. 2008, 283, 8868–8876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Ruiz, J.M.; Spínola, M.C.; Kirchsteiger, K.; Moreno, J.; Sahrawy, M.; Cejudo, F.J. Rice NTRC Is a High-Efficiency Redox System for Chloroplast Protection against Oxidative Damage. Plant Cell 2006, 18, 2356–2368. [Google Scholar] [CrossRef] [Green Version]
- Fodor, G.; Arnold, R.; Mohácsi, T.; Karle, I.; Flippen-Anderson, J. A new role for l-ascorbic acid: Michael donor to α,β-unsaturated carbonyl compounds. Tetrahedron 1983, 39, 2137–2145. [Google Scholar] [CrossRef]
- Schauenstein, E.; Taufer, M.; Esterbauer, H.; Kylianek, A.; Seelich, T. Über die Reaktion von Protein-SH-Gruppen mit 4-Hydroxypentenal. Mon. Chem. Chem. Mon. 1971, 102, 517–529. [Google Scholar] [CrossRef]
- Dennehy, M.K.; Richards, K.A.M.; Wernke, G.R.; Shyr, Y.; Liebler, D.C. Cytosolic and Nuclear Protein Targets of Thiol-Reactive Electrophiles. Chem. Res. Toxicol. 2006, 19, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hartman, H.; Syvanen, M.; Buchanan, B.B. Contrasting evolutionary histories of chloroplast thioredoxins f and m. Mol. Biol. Evol. 1990, 7, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Holmgren, A. Tryptophan Fluorescence Study of Conformational Transitions of the Oxidized and Reduced Form of Thioredoxin. J. Biol. Chem. 1972, 247, 1992–1998. [Google Scholar] [CrossRef]
- Chibani, K.; Saul, F.; Didierjean, C.; Rouhier, N.; Haouz, A. Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1. FEBS Lett. 2018, 592, 1030–1041. [Google Scholar] [CrossRef] [Green Version]
- Hayter, J.R.; Robertson, D.H.L.; Gaskell, S.J.; Beynon, R.J. Proteome Analysis of Intact Proteins in Complex Mixtures. Mol. Cell. Proteom. 2003, 2, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, J.; Muthuramalingam, M.; Dietz, K.-J. Mechanisms and dynamics in the thiol/disulfide redox regulatory network: Transmitters, sensors and targets. Curr. Opin. Plant Biol. 2012, 15, 261–268. [Google Scholar] [CrossRef]
- Wang, M.; Weiss, M.; Simonovic, M.; Haertinger, G.; Schrimpf, S.P.; Hengartner, M.O.; Von Mering, C. PaxDb, a Database of Protein Abundance Averages Across All Three Domains of Life*. Mol. Cell. Proteom. 2012, 11, 492–500. [Google Scholar] [CrossRef] [Green Version]
- Cheong, H.; Dos Santos, I.B.; Liu, W.; Gosse, H.N.; Park, S.-W. Cyclophilin 20–3 is positioned as a regulatory hub between light-dependent redox and 12-oxo-phytodienoic acid signaling. Plant Signal. Behav. 2017, 12, e1362520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Dos Santos, I.B.; Moye, A.; Park, S.-W. CYP20-3 deglutathionylates 2-CysPRX A and suppresses peroxide detoxification during heat stress. Life Sci. Alliance 2020, 3, e202000775. [Google Scholar] [CrossRef]
- LoPachin, R.M.; Barber, D.S.; Gavin, T. Molecular Mechanisms of the Conjugated α,β-Unsaturated Carbonyl Derivatives: Relevance to Neurotoxicity and Neurodegenerative Diseases. Toxicol. Sci. 2007, 104, 235–249. [Google Scholar] [CrossRef]
- Mueller, M.J.; Berger, S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry 2009, 70, 1511–1521. [Google Scholar] [CrossRef]
- Mano, J.; Nagata, M.; Okamura, S.; Shiraya, T.; Mitsui, T. Identification of Oxidatively Modified Proteins in Salt-Stressed Arabidopsis: A Carbonyl-Targeted Proteomics Approach. Plant Cell Physiol. 2014, 55, 1233–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löwe, J.; Dietz, K.; Gröger, H. From a Biosynthetic Pathway toward a Biocatalytic Process and Chemocatalytic Modifications: Three-Step Enzymatic Cascade to the Plant Metabolite cis -(+)-12-OPDA and Metathesis-Derived Products. Adv. Sci. 2020, 7, 1902973. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis Endomembrane System. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portoghese, P.; Kedziora, G.; Larson, D.; Bernard, B.; Hall, R. Reactivity of glutathione with α,β-unsaturated ketone flavouring substances. Food Chem. Toxicol. 1989, 27, 773–776. [Google Scholar] [CrossRef]
- Kossmann, J.; Sonnewald, U.; Willmitzer, L. Reduction of the chloroplastic fructose-1,6-bisphosphatase in transgenic potato plants impairs photosynthesis and plant growth. Plant J. 1994, 6, 637–650. [Google Scholar] [CrossRef]
- Michelet, L.; Zaffagnini, M.; Marchand, C.; Collin, V.; Decottignies, P.; Tsan, P.; Lancelin, J.-M.; Trost, P.; Miginiac-Maslow, M.; Noctor, G.; et al. Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 16478–16483. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.M.; Wang, S.; Telman, W.; Liebthal, M.; Schnitzer, H.; Viehhauser, A.; Sticht, C.; Delatorre, C.; Wirtz, M.; Hell, R.; et al. The redox-sensitive module of cyclophilin 20-3, 2-cysteine peroxiredoxin and cysteine synthase integrates sulfur metabolism and oxylipin signaling in the high light acclimation response. Plant J. 2017, 91, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- Lippuner, V.; Chou, I.; Scott, S.; Ettinger, W.; Theg, S.; Gasser, C. Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J. Biol. Chem. 1994, 269, 7863–7868. [Google Scholar] [CrossRef]
- Deng, W.; Chen, L.; Wood, D.W.; Metcalfe, T.; Liang, X.; Gordon, M.P.; Comai, L.; Nester, E.W. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl. Acad. Sci. USA 1998, 95, 7040–7045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Liu, W.; Sun, L. Role of cyclophilin A during coronavirus replication and the antiviral activities of its inhibitors. Chin. J. Biotechnol. 2020, 36, 605–611. [Google Scholar]
- Monte, I.; Kneeshaw, S.; Franco-Zorrilla, J.M.; Chini, A.; Zamarreño, A.M.; García-Mina, J.M.; Solano, R. An Ancient COI1-Independent Function for Reactive Electrophilic Oxylipins in Thermotolerance. Curr. Biol. 2020, 30, 962–971.e3. [Google Scholar] [CrossRef] [PubMed]
- Hochmal, A.K.; Zinzius, K.; Charoenwattanasatien, R.; Gäbelein, P.; Mutoh, R.; Tanaka, H.; Schulze, S.; Liu, G.; Scholz, M.; Nordhues, A.; et al. Calredoxin represents a novel type of calcium-dependent sensor-responder connected to redox regulation in the chloroplast. Nat. Commun. 2016, 7, 11847. [Google Scholar] [CrossRef] [PubMed]







| Sum Formula | Name | Charge at pH 7 | RBN | logP | TPSA | Physiological Relevance |
|---|---|---|---|---|---|---|
| C18H28O3 | 12-Oxophytodienoic acid, 12-OPDA | −1 | 11 | 4.58 | 54.37 | 2-cyclopentenone prostaglandin analogue, thiol metabolism, stress response, JA precursor [9] |
| C20H32O3 | 12-OPDA-ethylester, 12-OPDA-Et | 0 | 11 | 5.27 | 43.38 | Ethylester of 12-OPDA, unknown |
| C5H6O | 2-Cyclopentenone, CP | 0 | 0 | 0.40 | 17.07 | Structural subunit of 12-OPDA and other CP-derivatives |
| C4H6O | Methyl vinyl ketone, MVK | 0 | 1 | 0.50 | 17.07 | Isoprene and lipid-derived stress-related oxidation product [4,5] |
| C5H8O | Cyclopentanone, CPa | 0 | 0 | 0.89 | 17.07 | Structural subunit of MJ and various PGs |
| C6H10O | Trans-2-hexenal, HEX | 0 | 3 | 2.33 | 17.07 | Green leaf volatile, stress response [3] |
| C9H16O2 | 4-Hydroxynonenal, 4-HNE | 0 | 6 | 2.46 | 37.30 | Lipid-derived stress-related oxidation product [10] |
| C15H20O4 | Abscisic acid, ABA | −1 | 3 | 2.10 | 74.60 | Isoprenoid-derived 2-cyclohexenone compound, sulfur and thiol metabolism, leaf abscission, stress response, germination [6,8] |
| C20H28O3 | 15-Deoxy-Δ12,14-prostaglandin J2, 15dPGJ2 | −1 | 11 | 5.21 | 54.37 | Mammalian 2-cyclopentenone prostaglandin, thiol metabolism, stress response [11,12] |
| C20H30O4 | Prostaglandin J2, PGJ2 | −1 | 12 | 3.76 | 74.60 | Mammalian 2-cyclopentenone prostaglandin, thiol metabolism, stress response [12] |
| Sample | [ligand]/[protein] | λEm-max [nm] | Fluorescence intensity [r. U] | n |
|---|---|---|---|---|
| TRX-f1red | 0 | 342.2 ± 0.3 | 73.3 ± 4.4 | 3 |
| TRX-f1red + 12-OPDA | 9.6 | 341.3 ± 0.4 | 64.6 ± 2.9 | 2 |
| TRX-f1red + 12-OPDA | 19.2 | 341.7 ± 0.3 | 60.1 ± 3.2 * | 3 |
| TRX-f1untreated | 0 | 341.3 ± 0.8 | 82.2 ± 0.6 | 3 |
| TRX-f1untreated + 12-OPDA | 18.4 | 342.2 ± 0.3 | 78.6 ± 1.4 * | 3 |
| TRX-m1red | 0 | 327.0 ± 0.4 | 81.6 ± 1.8 | 4 |
| TRX-m1red + 12-OPDA | 18.4 | 328.5 ± 0.7 | 73.0 ± 1.4 * | 4 |
| TRX-m1untreated | 0 | 315.3 ± 0.3 | 74.3 ± 2.2 | 4 |
| TRX-m1untreated + 12-OPDA | 18.4 | 315.1 ± 0.5 | 68.6 ± 1.3 * | 4 |
| 2-CysPRXuntreated | 0 | 338.3 ± 0.3 | 86.9 ± 0.9 | 6 |
| 2-CysPRXuntreated + 12-OPDA | 18.4 | 339.3 ± 0.3 * | 83.1 ± 0.6 * | 4 |
| 2-CysPRXred | 0 | 335.2 ± 0.3 | 97.1 ± 1.5 | 3 |
| 2-CysPRXred+12-OPDA | 18.4 | 336.5 ± 0.4 * | 92.5 ± 0.9 * | 4 |
| Additive | Change in Fluorescence Intensity [r. U] at λEm-max [nm] Relative to Control | λEm-max [nm] | |
|---|---|---|---|
| Treatment | Control | ||
| 12-OPDA | −2.52 ± 0.50 * | 331.63 ± 0.95 | 330.13 ± 0.25 |
| MVK | −1.48 ± 0.48 * | 330.80 ± 0.57 | 330.25 ± 0.87 |
| CP | −0.66 ± 0.7 | 330.33 ± 0.29 | 330.00 ± 0.00 |
| CPa | −0.24 ± 0.5 | 330.13 ± 0.25 | 330.38 ± 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maynard, D.; Viehhauser, A.; Knieper, M.; Dreyer, A.; Manea, G.; Telman, W.; Butter, F.; Chibani, K.; Scheibe, R.; Dietz, K.-J. The In Vitro Interaction of 12-Oxophytodienoic Acid and Related Conjugated Carbonyl Compounds with Thiol Antioxidants. Biomolecules 2021, 11, 457. https://doi.org/10.3390/biom11030457
Maynard D, Viehhauser A, Knieper M, Dreyer A, Manea G, Telman W, Butter F, Chibani K, Scheibe R, Dietz K-J. The In Vitro Interaction of 12-Oxophytodienoic Acid and Related Conjugated Carbonyl Compounds with Thiol Antioxidants. Biomolecules. 2021; 11(3):457. https://doi.org/10.3390/biom11030457
Chicago/Turabian StyleMaynard, Daniel, Andrea Viehhauser, Madita Knieper, Anna Dreyer, Ghamdan Manea, Wilena Telman, Falk Butter, Kamel Chibani, Renate Scheibe, and Karl-Josef Dietz. 2021. "The In Vitro Interaction of 12-Oxophytodienoic Acid and Related Conjugated Carbonyl Compounds with Thiol Antioxidants" Biomolecules 11, no. 3: 457. https://doi.org/10.3390/biom11030457