Macromolecular Crowding Increases the Affinity of the PHD of ING4 for the Histone H3K4me3 Mark
Abstract
:1. Introduction
2. Materials and Methods
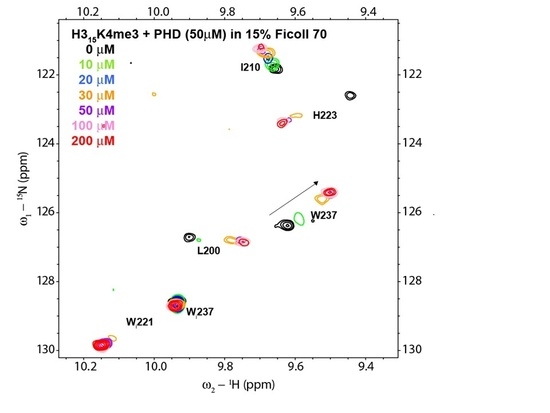
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- de la Cruz, X.; Lois, S.; Sanchez-Molina, S.; Martinez-Balbas, M.A. Do protein motifs read the histone code? Bioessays 2005, 27, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Santos-Rosa, H.; Caldas, C. Chromatin modifier enzymes, the histone code and cancer. Eur. J. Cancer Prev. 2005, 41, 2381–2402. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nightingale, K.P.; O’Neill, L.P.; Turner, B.M. Histone modifications: Signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev. 2006, 16, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Lall, S. Primers on chromatin. Nat. Struct. Mol. Biol. 2007, 14, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.D.; Li, H.; Ruthenburg, A.J.; Allis, C.D.; Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007, 14, 1025–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallen, G.; Riabowol, K. Keep-ING balance: tumor suppression by epigenetic regulation. FEBS Lett. 2014, 588, 2728–2742. [Google Scholar] [CrossRef] [Green Version]
- Peña, P.V.; Davrazou, F.; Shi, X.; Walter, K.L.; Verkhusha, V.V.; Gozani, O.; Zhao, R.; Kutateladze, T.G. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 2006, 442, 100–103. [Google Scholar] [CrossRef]
- Palacios, A.; Palacios, A.; Moreno, A.; Oliveira, B.L.; Rivera, T.; Prieto, J.; Garcia, P.; Fernandez-Fernandez, M.R.; Bernado, P.; Palmero, I.; et al. The dimeric structure and the bivalent recognition of H3K4me3 by the tumor suppressor ING4 suggests a mechanism for enhanced targeting of the HBO1 complex to chromatin. J. Mol. Biol. 2010, 396, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Culurgioni, S.; Munoz, I.G.; Moreno, A.; Palacios, A.; Villate, M.; Palmero, I.; Montoya, G.; Blanco, F.J. The crystal structure of the inhibitor of growth 4 (ING4) dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. J. Biol. Chem. 2012, 287, 10876–10884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ormaza, G.; Medagli, B.; Ibanez de Opakua, A.; Rodriguez, J.A.; Merino, N.; Villate, M.; Onesti, S.; Blanco, F.J. The tumor suppressor inhibitor of growth 4 binds double-stranded DNA through its disordered central region. FEBS Lett. 2017, 591, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Palacios, A.; Garcia, P.; Padro, D.; Lopez-Hernandez, E.; Martin, I.; Blanco, F.J. Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4. FEBS Lett. 2006, 205 580, 6903–6908. [Google Scholar] [CrossRef] [Green Version]
- Palacios, A.; Munoz, I.G.; Pantoja-Uceda, D.; Marcaida, M.J.; Torres, D.; Martin-Garcia, J.M.; Luque, I.; Montoya, G.; Blanco, F.J. Molecular basis of histone H3K4me3 recognition by ING4. J. Biol. Chem. 2008, 283, 15956–15964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minton, A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000, 10, 34–39. [Google Scholar] [CrossRef]
- Rivas, G.; Minton, A.P. Macromolecular Crowding In Vitro, In Vivo, and In Between. Trends Biochem. Sci. 2016, 41, 970–981. [Google Scholar] [CrossRef] [Green Version]
- Luby-Phelps, K.; Castle, P.E.; Taylor, D.L.; Lanni, F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc. Natl. Acad. Sci. USA 1987, 84, 4910–4913. [Google Scholar] [CrossRef] [Green Version]
- Monterroso, B.; Minton, A.P. Effect of high concentration of inert cosolutes on the refolding of an enzyme: carbonic anhydrase B in sucrose and ficoll 70. J. Biol. Chem. 2007, 282, 33452–33458. [Google Scholar] [CrossRef] [Green Version]
- Venturoli, D.; Rippe, B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am. J. Physiol. Renal Physiol. 2005, 288, F605–F613. [Google Scholar] [CrossRef]
- Ormaza, G.; Rodriguez, J.A.; Ibanez de Opakua, A.; Merino, N.; Villate, M.; Gorrono, I.; Rabano, M.; Palmero, I.; Vilaseca, M.; Kypta, R.; et al. The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark. J. Mol. Biol. 2019, 431, 2298–2319. [Google Scholar] [CrossRef]
- Rivas, G.; Minton, A.P. Toward an understanding of biochemical equilibria within living cells. Biophys. Rev. 2018, 10, 241–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, X.; Nguyen, A.W.; Jabaiah, A.; Sheff, M.A.; Thorn, K.S.; Daugherty, P.S. Intracellular protein interaction mapping with FRET hybrids. Proc. Natl. Acad. Sci. USA 2006, 103, 18458–18463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallen, G.; Riabowol, K. DisorderING promotes epigenetic order. FEBS Lett. 2017, 591, 257–259. [Google Scholar] [CrossRef] [Green Version]
- Avvakumov, N.; Lalonde, M.E.; Saksouk, N.; Paquet, E.; Glass, K.C.; Landry, A.J.; Doyon, Y.; Cayrou, C.; Robitaille, G.A.; Richard, D.E.; et al. Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell. Biol. 2011, 32, 689–703. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palacios, A.; Blanco, F.J. Macromolecular Crowding Increases the Affinity of the PHD of ING4 for the Histone H3K4me3 Mark. Biomolecules 2020, 10, 234. https://doi.org/10.3390/biom10020234
Palacios A, Blanco FJ. Macromolecular Crowding Increases the Affinity of the PHD of ING4 for the Histone H3K4me3 Mark. Biomolecules. 2020; 10(2):234. https://doi.org/10.3390/biom10020234
Chicago/Turabian StylePalacios, Alicia, and Francisco J Blanco. 2020. "Macromolecular Crowding Increases the Affinity of the PHD of ING4 for the Histone H3K4me3 Mark" Biomolecules 10, no. 2: 234. https://doi.org/10.3390/biom10020234