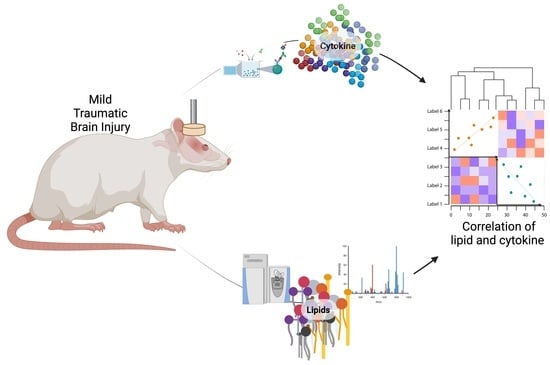
Integrative Analysis of Cytokine and Lipidomics Datasets Following Mild Traumatic Brain Injury in Rats
Abstract
:1. Introduction
2. Material and Methods
2.1. Injury Protocol
2.2. Sample Collection and Preparation
2.3. Sample Analysis with Ultra-High Performance Liquid Chromatography–Mass Spectrometry (UPLC-MS)
2.4. UPLC-MS Data Processing
2.5. Luminex Methods
2.6. Statistical Analysis
3. Experimental Design
4. Results
4.1. Brain Lipidomic Changes after mTBI
4.1.1. Statistical Analysis of Lipid Content in Brain Cortices Detected by LC-MS
4.1.2. Minimization of Experimental Batch Effects in the Brain Cortices Dataset
4.1.3. Lipid Annotation
4.2. Evaluation of Inflammation Markers after mTBI
4.3. Integration of Lipidomics and Cytokine Datasets
5. Discussion
5.1. Combining Lipidomic and Cytokine Data
5.2. Limitations, Challenges, and Opportunities
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dewan, M.C.; Rattani, A.; Gupta, S.; Baticulon, R.E.; Hung, Y.C.; Punchak, M.; Agrawal, A.; Adeleye, A.O.; Shrime, M.G.; Rubiano, A.M.; et al. Estimating the global incidence of traumatic brain injury. J. Neurosurg. 2018, 130, 1080–1097. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Health and Human Services. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations, and Deaths, 2002–2006; United States Department of Health and Human Services: Washington, DC, USA, 2010.
- Alkhaibary, A.; Alshalawi, A.; Althaqafi, R.M.M.; Alghuraybi, A.A.; Basalamah, A.; Shammaa, A.M.; Altalhy, A.A.; Abdelrahman, T.M. Traumatic Brain Injury: A Perspective on the Silent Epidemic. Cureus 2021, 13, e15318. [Google Scholar] [CrossRef] [PubMed]
- Coburn, K. Traumatic brain injury: The silent epidemic. AACN Clin. Issues Crit. Care Nurs. 1992, 3, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rodriguez, A.; Egea-Guerrero, J.J.; Murillo-Cabezas, F.; Carrillo-Vico, A. Oxidative stress in traumatic brain injury. Curr. Med. Chem. 2014, 21, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Baracaldo-Santamaría, D.; Ariza-Salamanca, D.F.; Corrales-Hernández, M.G.; Pachón-Londoño, M.J.; Hernandez-Duarte, I.; Calderon-Ospina, C.-A. Revisiting Excitotoxicity in Traumatic Brain Injury: From Bench to Bedside. Pharmaceutics 2022, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.; Bejjani, F.; Raad, M.; Abou-El-Hassan, H.; Mantash, S.; Nokkari, A.; Ramadan, N.; Kassem, N.; Mondello, S.; Hamade, E.; et al. Traumatic Brain Injury and Blood-Brain Barrier Cross-Talk. CNS Neurol. Disord. Drug Targets 2016, 15, 1030–1044. [Google Scholar] [CrossRef]
- LaPlaca, M.C.; Prado, G.R. Neural mechanobiology and neuronal vulnerability to traumatic loading. J. Biomech. 2010, 43, 71–78. [Google Scholar] [CrossRef]
- Raghupathi, R. Cell death mechanisms following traumatic brain injury. Brain Pathol. 2004, 14, 215–222. [Google Scholar] [CrossRef]
- Simon, D.W.; McGeachy, M.J.; Bayır, H.; Clark, R.S.; Loane, D.J.; Kochanek, P.M. The far-reaching scope of neuroinflammation after traumatic brain injury. Nat. Rev. Neurol. 2017, 13, 171–191. [Google Scholar] [CrossRef]
- Nessel, I.; Michael-Titus, A.T. Lipid profiling of brain tissue and blood after traumatic brain injury: A review of human and experimental studies. Semin. Cell Dev. Biol. 2021, 112, 145–156. [Google Scholar] [CrossRef]
- Sparvero, L.J.; Amoscato, A.A.; Kochanek, P.M.; Pitt, B.R.; Kagan, V.E.; Bayir, H. Mass-spectrometry based oxidative lipidomics and lipid imaging: Applications in traumatic brain injury. J. Neurochem. 2010, 115, 1322–1336. [Google Scholar] [CrossRef]
- Leuti, A.; Fazio, D.; Fava, M.; Piccoli, A.; Oddi, S.; Maccarrone, M. Bioactive lipids, inflammation and chronic diseases. Adv. Drug Deliv. Rev. 2020, 159, 133–169. [Google Scholar] [CrossRef]
- Poblete, R.A.; Arenas, M.; Sanossian, N.; Freeman, W.D.; Louie, S.G. The role of bioactive lipids in attenuating the neuroinflammatory cascade in traumatic brain injury. Ann. Clin. Transl. Neurol. 2020, 7, 2524–2534. [Google Scholar] [CrossRef]
- Shohami, E.; Shapira, Y.; Yadid, G.; Reisfeld, N.; Yedgar, S. Brain phospholipase A2 is activated after experimental closed head injury in the rat. J. Neurochem. 1989, 53, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.C.; Liao, Y.E.; Yang, L.Y.; Wang, J.Y.; Tweedie, D.; Karnati, H.K.; Greig, N.H.; Wang, J.Y. Neuroinflammation in animal models of traumatic brain injury. J. Neurosci. Methods 2016, 272, 38–49. [Google Scholar] [CrossRef]
- Juengst, S.B.; Kumar, R.G.; Failla, M.D.; Goyal, A.; Wagner, A.K. Acute inflammatory biomarker profiles predict depression risk following moderate to severe traumatic brain injury. J. Head. Trauma. Rehabil. 2015, 30, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Morganti-Kossmann, M.C.; Semple, B.D.; Hellewell, S.C.; Bye, N.; Ziebell, J.M. The complexity of neuroinflammation consequent to traumatic brain injury: From research evidence to potential treatments. Acta Neuropathol. 2019, 137, 731–755. [Google Scholar] [CrossRef]
- Ozaki, K.; Leonard, W.J. Cytokine and cytokine receptor pleiotropy and redundancy. J. Biol. Chem. 2002, 277, 29355–29358. [Google Scholar] [CrossRef]
- Ziebell, J.M.; Morganti-Kossmann, M.C. Involvement of pro- and anti-inflammatory cytokines and chemokines in the pathophysiology of traumatic brain injury. Neurother. J. Am. Soc. Exp. NeuroTherapeutics 2010, 7, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.G.; Geddes, R.I.; Sribnick, E.A. Recent developments in clinical trials for the treatment of traumatic brain injury. Handb. Clin. Neurol. 2015, 127, 433–451. [Google Scholar] [CrossRef]
- Pulliam, A.N.; Gier, E.C.; Gaul, D.A.; Moore, S.G.; Fernandez, F.M.; LaPlaca, M.C. Comparing Brain and Blood Lipidome Changes following Single and Repetitive Mild Traumatic Brain Injury in Rats. ACS Chem. Neurosci. 2024, 15, 300–314. [Google Scholar] [CrossRef]
- Bodnar, C.N.; Roberts, K.N.; Higgins, E.K.; Bachstetter, A.D. A Systematic Review of Closed Head Injury Models of Mild Traumatic Brain Injury in Mice and Rats. J. Neurotrauma 2019, 36, 1683–1706. [Google Scholar] [CrossRef]
- Creed, J.A.; DiLeonardi, A.M.; Fox, D.P.; Tessler, A.R.; Raghupathi, R. Concussive brain trauma in the mouse results in acute cognitive deficits and sustained impairment of axonal function. J. Neurotrauma 2011, 28, 547–563. [Google Scholar] [CrossRef]
- Hylin, M.J.; Orsi, S.A.; Rozas, N.S.; Hill, J.L.; Zhao, J.; Redell, J.B.; Moore, A.N.; Dash, P.K. Repeated mild closed head injury impairs short-term visuospatial memory and complex learning. J. Neurotrauma 2013, 30, 716–726. [Google Scholar] [CrossRef]
- Statler, K.D.; Kochanek, P.M.; Dixon, C.E.; Alexander, H.L.; Warner, D.S.; Clark, R.S.; Wisniewski, S.R.; Graham, S.H.; Jenkins, L.W.; Marion, D.W.; et al. Isoflurane improves long-term neurologic outcome versus fentanyl after traumatic brain injury in rats. J. Neurotrauma 2000, 17, 1179–1189. [Google Scholar] [CrossRef]
- Statler, K.D.; Alexander, H.; Vagni, V.; Holubkov, R.; Dixon, C.E.; Clark, R.S.; Jenkins, L.; Kochanek, P.M. Isoflurane exerts neuroprotective actions at or near the time of severe traumatic brain injury. Brain Res. 2006, 1076, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Yurdakoc, A.; Gunday, I.; Memis, D. Effects of halothane, isoflurane, and sevoflurane on lipid peroxidation following experimental closed head trauma in rats. Acta Anaesthesiol. Scand. 2008, 52, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Le Freche, H.; Brouillette, J.; Fernandez-Gomez, F.J.; Patin, P.; Caillierez, R.; Zommer, N.; Sergeant, N.; Buee-Scherrer, V.; Lebuffe, G.; Blum, D.; et al. Tau phosphorylation and sevoflurane anesthesia: An association to postoperative cognitive impairment. Anesthesiology 2012, 116, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.S.; Muresanu, D.F.; Nozari, A.; Castellani, R.J.; Dey, P.K.; Wiklund, L.; Sharma, A. Anesthetics influence concussive head injury induced blood-brain barrier breakdown, brain edema formation, cerebral blood flow, serotonin levels, brain pathology and functional outcome. Int. Rev. Neurobiol. 2019, 146, 45–81. [Google Scholar] [CrossRef] [PubMed]
- Wojnarowicz, M.W.; Fisher, A.M.; Minaeva, O.; Goldstein, L.E. Considerations for Experimental Animal Models of Concussion, Traumatic Brain Injury, and Chronic Traumatic Encephalopathy—These Matters Matter. Front. Neurol. 2017, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Ahlers, S.T.; Vasserman-Stokes, E.; Shaughness, M.C.; Hall, A.A.; Shear, D.A.; Chavko, M.; McCarron, R.M.; Stone, J.R. Assessment of the effects of acute and repeated exposure to blast overpressure in rodents: Toward a greater understanding of blast and the potential ramifications for injury in humans exposed to blast. Front. Neurol. 2012, 3, 32. [Google Scholar] [CrossRef]
- Petraglia, A.L.; Plog, B.A.; Dayawansa, S.; Chen, M.; Dashnaw, M.L.; Czerniecka, K.; Walker, C.T.; Viterise, T.; Hyrien, O.; Iliff, J.J.; et al. The spectrum of neurobehavioral sequelae after repetitive mild traumatic brain injury: A novel mouse model of chronic traumatic encephalopathy. J. Neurotrauma 2014, 31, 1211–1224. [Google Scholar] [CrossRef]
- Bolouri, H.; Zetterberg, H. Animal Models for Concussion: Molecular and Cognitive Assessments-Relevance to Sport and Military Concussions. In Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects; Kobeissy, F.H., Ed.; Frontiers in Neuroengineering: Boca Raton, FL, USA, 2015. [Google Scholar]
- Hoogenboom, W.S.; Branch, C.A.; Lipton, M.L. Animal models of closed-skull, repetitive mild traumatic brain injury. Pharmacol. Ther. 2019, 198, 109–122. [Google Scholar] [CrossRef]
- Shultz, S.R.; McDonald, S.J.; Vonder Haar, C.; Meconi, A.; Vink, R.; van Donkelaar, P.; Taneja, C.; Iverson, G.L.; Christie, B.R. The potential for animal models to provide insight into mild traumatic brain injury: Translational challenges and strategies. Neurosci. Biobehav. Rev. 2017, 76, 396–414. [Google Scholar] [CrossRef] [PubMed]
- Bignall, K.E. Ontogeny of levels of neural organization: The righting reflex as a model. Exp. Neurol. 1974, 42, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Franks, N.P. General anaesthesia: From molecular targets to neuronal pathways of sleep and arousal. Nat. Rev. Neurosci. 2008, 9, 370–386. [Google Scholar] [CrossRef] [PubMed]
- Hallam, T.M.; Floyd, C.L.; Folkerts, M.M.; Lee, L.L.; Gong, Q.Z.; Lyeth, B.G.; Muizelaar, J.P.; Berman, R.F. Comparison of behavioral deficits and acute neuronal degeneration in rat lateral fluid percussion and weight-drop brain injury models. J. Neurotrauma 2004, 21, 521–539. [Google Scholar] [CrossRef]
- Berman, R.; Spencer, H.; Boese, M.; Kim, S.; Radford, K.; Choi, K. Loss of Consciousness and Righting Reflex Following Traumatic Brain Injury: Predictors of Post-Injury Symptom Development (A Narrative Review). Brain Sci. 2023, 13, 750. [Google Scholar] [CrossRef]
- Gier, E.C.; Pulliam, A.N.; Gaul, D.A.; Moore, S.G.; LaPlaca, M.C.; Fernández, F.M. Lipidome Alterations following Mild Traumatic Brain Injury in the Rat. Metabolites 2022, 12, 150. [Google Scholar] [CrossRef]
- Sankar, S.B.; Pybus, A.F.; Liew, A.; Sanders, B.; Shah, K.J.; Wood, L.B.; Buckley, E.M. Low cerebral blood flow is a non-invasive biomarker of neuroinflammation after repetitive mild traumatic brain injury. Neurobiol. Dis. 2019, 124, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Sarkaki, A.R.; Khaksari Haddad, M.; Soltani, Z.; Shahrokhi, N.; Mahmoodi, M. Time- and dose-dependent neuroprotective effects of sex steroid hormones on inflammatory cytokines after a traumatic brain injury. J. Neurotrauma 2013, 30, 47–54. [Google Scholar] [CrossRef]
- He, J.; Evans, C.O.; Hoffman, S.W.; Oyesiku, N.M.; Stein, D.G. Progesterone and allopregnanolone reduce inflammatory cytokines after traumatic brain injury. Exp. Neurol. 2004, 189, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, A.C.; Kim, H.; Salcedo, E.; Yonchek, J.C.; Rodgers, K.M.; Orfila, J.E.; Dietz, R.M.; Quillinan, N.; Traystman, R.J.; Herson, P.S. Endogenous Sex Steroids Dampen Neuroinflammation and Improve Outcome of Traumatic Brain Injury in Mice. J. Mol. Neurosci. 2018, 64, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Homayoun, P.; Parkins, N.E.; Soblosky, J.; Carey, M.E.; Rodriguez de Turco, E.B.; Bazan, N.G. Cortical impact injury in rats promotes a rapid and sustained increase in polyunsaturated free fatty acids and diacylglycerols. Neurochem. Res. 2000, 25, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Muller, L.; Jackson, S.N.; Post, J.; Baldwin, K.; Hoffer, B.; Balaban, C.D.; Barbacci, D.; Schultz, J.A.; Gouty, S.; et al. Mass spectrometry imaging of rat brain lipid profile changes over time following traumatic brain injury. J. Neurosci. Methods 2016, 272, 19–32. [Google Scholar] [CrossRef]
- Ojo, J.O.; Algamal, M.; Leary, P.; Abdullah, L.; Mouzon, B.; Evans, J.E.; Mullan, M.; Crawford, F. Disruption in Brain Phospholipid Content in a Humanized Tau Transgenic Model Following Repetitive Mild Traumatic Brain Injury. Front. Neurosci. 2018, 12, 893. [Google Scholar] [CrossRef] [PubMed]
- Tweedie, D.; Karnati, H.K.; Mullins, R.; Pick, C.G.; Hoffer, B.J.; Goetzl, E.J.; Kapogiannis, D.; Greig, N.H. Time-dependent cytokine and chemokine changes in mouse cerebral cortex following a mild traumatic brain injury. eLife 2020, 9, e55827. [Google Scholar] [CrossRef] [PubMed]
- Marion-Letellier, R.; Savoye, G.; Ghosh, S. Polyunsaturated fatty acids and inflammation. IUBMB Life 2015, 67, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.L.; McDonald, D.A.; Borum, P.R. Acylcarnitines: Role in brain. Prog. Lipid Res. 2010, 49, 61–75. [Google Scholar] [CrossRef]
- Falchetto, S.; Kato, G.; Provini, L. The action of carnitines on cortical neurons. Can. J. Physiol. Pharmacol. 1971, 49, 1–7. [Google Scholar] [CrossRef]
- Spagnoli, A.; Lucca, U.; Menasce, G.; Bandera, L.; Cizza, G.; Forloni, G.; Tettamanti, M.; Frattura, L.; Tiraboschi, P.; Comelli, M.; et al. Long-term acetyl-L-carnitine treatment in Alzheimer’s disease. Neurology 1991, 41, 1726–1732. [Google Scholar] [CrossRef]
- Forloni, G.; Angeretti, N.; Smiroldo, S. Neuroprotective activity of acetyl-L-carnitine: Studies in vitro. J. Neurosci. Res. 1994, 37, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Scafidi, S.; Racz, J.; Hazelton, J.; McKenna, M.C.; Fiskum, G. Neuroprotection by acetyl-L-carnitine after traumatic injury to the immature rat brain. Dev. Neurosci. 2010, 32, 480–487. [Google Scholar] [CrossRef]
- Bazinet, R.P.; Laye, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Dhillon, H.S.; Donaldson, D.; Dempsey, R.J.; Prasad, M.R. Regional levels of free fatty acids and Evans blue extravasation after experimental brain injury. J. Neurotrauma 1994, 11, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Strum, J.C.; Bell, R.M. Lipid biochemistry: Functions of glycerolipids and sphingolipids in cellular signaling. FASEB J. 1997, 11, 45–50. [Google Scholar] [CrossRef]
- Voelker, D.R. Glycerolipid Structure, Function, and Synthesis in Eukaryotes. In Encyclopedia of Biological Chemistry, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 412–418. [Google Scholar] [CrossRef]
- Ogasawara, D.; Deng, H.; Viader, A.; Baggelaar, M.P.; Breman, A.; den Dulk, H.; van den Nieuwendijk, A.M.; Soethoudt, M.; van der Wel, T.; Zhou, J.; et al. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc. Natl. Acad. Sci. USA 2016, 113, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A.; Farr, S.A.; Salameh, T.S.; Niehoff, M.L.; Rhea, E.M.; Morley, J.E.; Hanson, A.J.; Hansen, K.M.; Craft, S. Triglycerides cross the blood-brain barrier and induce central leptin and insulin receptor resistance. Int. J. Obes. 2018, 42, 391–397. [Google Scholar] [CrossRef]
- Xie, Y.; Li, J.; Kang, R.; Tang, D. Interplay Between Lipid Metabolism and Autophagy. Front. Cell Dev. Biol. 2020, 8, 431. [Google Scholar] [CrossRef]
- Saito, T.; Kuma, A.; Sugiura, Y.; Ichimura, Y.; Obata, M.; Kitamura, H.; Okuda, S.; Lee, H.C.; Ikeda, K.; Kanegae, Y.; et al. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat. Commun. 2019, 10, 1567. [Google Scholar] [CrossRef]
- Lim, J.; Yang, E.J.; Chang, J.H. Upregulation of TNF-α by Triglycerides is Mediated by MEK1 Activation in Jurkat T Cells. Biomed. Sci. Lett. 2018, 24, 213–220. [Google Scholar] [CrossRef]
- Dai, Y.; Tang, H.; Pang, S. The Crucial Roles of Phospholipids in Aging and Lifespan Regulation. Front. Physiol. 2021, 12, 775648. [Google Scholar] [CrossRef]
- Abdullah, L.; Evans, J.E.; Ferguson, S.; Mouzon, B.; Montague, H.; Reed, J.; Crynen, G.; Emmerich, T.; Crocker, M.; Pelot, R.; et al. Lipidomic analyses identify injury-specific phospholipid changes 3 mo after traumatic brain injury. FASEB J. 2014, 28, 5311–5321. [Google Scholar] [CrossRef]
- Yang, B.; Fritsche, K.L.; Beversdorf, D.Q.; Gu, Z.; Lee, J.C.; Folk, W.R.; Greenlief, C.M.; Sun, G.Y. Yin-Yang Mechanisms Regulating Lipid Peroxidation of Docosahexaenoic Acid and Arachidonic Acid in the Central Nervous System. Front. Neurol. 2019, 10, 642. [Google Scholar] [CrossRef]
- Chao, H.; Lin, C.; Zuo, Q.; Liu, Y.; Xiao, M.; Xu, X.; Li, Z.; Bao, Z.; Chen, H.; You, Y.; et al. Cardiolipin-Dependent Mitophagy Guides Outcome after Traumatic Brain Injury. J. Neurosci. 2019, 39, 1930–1943. [Google Scholar] [CrossRef]
- Pointer, C.B.; Klegeris, A. Cardiolipin in Central Nervous System Physiology and Pathology. Cell. Mol. Neurobiol. 2017, 37, 1161–1172. [Google Scholar] [CrossRef]
- Anthonymuthu, T.S.; Kenny, E.M.; Hier, Z.E.; Clark, R.S.B.; Kochanek, P.M.; Kagan, V.E.; Bayir, H. Detection of brain specific cardiolipins in plasma after experimental pediatric head injury. Exp. Neurol. 2019, 316, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.B.; Faergeman, N.J. Sphingolipids: Membrane microdomains in brain development, function and neurological diseases. Open Biol. 2017, 7, 170069. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H., Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B.; Hannun, Y.A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer 2004, 4, 604–616. [Google Scholar] [CrossRef]
- Novgorodov, S.A.; Riley, C.L.; Yu, J.; Borg, K.T.; Hannun, Y.A.; Proia, R.L.; Kindy, M.S.; Gudz, T.I. Essential roles of neutral ceramidase and sphingosine in mitochondrial dysfunction due to traumatic brain injury. J. Biol. Chem. 2014, 289, 13142–13154. [Google Scholar] [CrossRef]
- Contrepois, K.; Liang, L.; Snyder, M. Can Metabolic Profiles Be Used as a Phenotypic Readout of the Genome to Enhance Precision Medicine? Clin. Chem. 2016, 62, 676–678. [Google Scholar] [CrossRef]
- Contrepois, K.; Mahmoudi, S.; Ubhi, B.K.; Papsdorf, K.; Hornburg, D.; Brunet, A.; Snyder, M. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep. 2018, 8, 17747. [Google Scholar] [CrossRef]
- Contrepois, K.; Jiang, L.; Snyder, M. Optimized Analytical Procedures for the Untargeted Metabolomic Profiling of Human Urine and Plasma by Combining Hydrophilic Interaction (HILIC) and Reverse-Phase Liquid Chromatography (RPLC)-Mass Spectrometry. Mol. Cell Proteom. 2015, 14, 1684–1695. [Google Scholar] [CrossRef]
- Ghosh, T.; Philtron, D.; Zhang, W.; Kechris, K.; Ghosh, D. Reproducibility of mass spectrometry based metabolomics data. BMC Bioinform. 2021, 22, 423. [Google Scholar] [CrossRef]
- Christians, U.; Klawitter, J.; Hornberger, A.; Klawitter, J. How unbiased is non-targeted metabolomics and is targeted pathway screening the solution? Curr. Pharm. Biotechnol. 2011, 12, 1053–1066. [Google Scholar] [CrossRef]
- Dill, A.L.; Eberlin, L.S.; Costa, A.B.; Ifa, D.R.; Cooks, R.G. Data quality in tissue analysis using desorption electrospray ionization. Anal. Bioanal. Chem. 2011, 401, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Shabihkhani, M.; Lucey, G.M.; Wei, B.; Mareninov, S.; Lou, J.J.; Vinters, H.V.; Singer, E.J.; Cloughesy, T.F.; Yong, W.H. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 2014, 47, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.H.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Oytam, Y.; Sobhanmanesh, F.; Duesing, K.; Bowden, J.C.; Osmond-McLeod, M.; Ross, J. Risk-conscious correction of batch effects: Maximising information extraction from high-throughput genomic datasets. BMC Bioinform. 2016, 17, 332. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ngoy, S.; Sheth, S.A.; Swanson, R.A.; Rhee, E.P.; Liao, R.; Clish, C.B.; Mootha, V.K.; Nilsson, R. A systematic survey of lipids across mouse tissues. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E854–E868. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Conroy, M.J.; Andrews, R.M.; Andrews, S.; Cockayne, L.; Dennis, E.A.; Fahy, E.; Gaud, C.; Griffiths, W.J.; Jukes, G.; Kolchin, M.; et al. LIPID MAPS: Update to databases and tools for the lipidomics community. Nucleic Acids Res. 2024, 52, D1677–D1682. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Shen, P.C.; Liu, H.C.; Cho, Y.C.; Hsu, M.K.; Lin, I.C.; Chen, F.H.; Yang, J.C.; Ma, W.L.; Cheng, W.C. LipidSig: A web-based tool for lipidomic data analysis. Nucleic Acids Res. 2021, 49, W336–W345. [Google Scholar] [CrossRef]
- Acevedo, A.; Durán, C.; Ciucci, S.; Gerl, M.; Cannistraci, C.V. LIPEA: Lipid pathway enrichment analysis. arXiv 2018, arXiv:274969. [Google Scholar]
- Signoretti, S.; Di Pietro, V.; Vagnozzi, R.; Lazzarino, G.; Amorini, A.M.; Belli, A.; D’Urso, S.; Tavazzi, B. Transient alterations of creatine, creatine phosphate, N-acetylaspartate and high-energy phosphates after mild traumatic brain injury in the rat. Mol. Cell Biochem. 2010, 333, 269–277. [Google Scholar] [CrossRef]
- Poblete, R.A.; Arenas, M.; Sanossian, N.; Hong, Y.K.; Freeman, W.D.; Lyden, P.D.; Louie, S.G. Pro-resolving lipid mediators in traumatic brain injury: Emerging concepts and translational approach. Am. J. Transl. Res. 2022, 14, 1482–1494. [Google Scholar]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulliam, A.N.; Pybus, A.F.; Gaul, D.A.; Moore, S.G.; Wood, L.B.; Fernández, F.M.; LaPlaca, M.C. Integrative Analysis of Cytokine and Lipidomics Datasets Following Mild Traumatic Brain Injury in Rats. Metabolites 2024, 14, 133. https://doi.org/10.3390/metabo14030133
Pulliam AN, Pybus AF, Gaul DA, Moore SG, Wood LB, Fernández FM, LaPlaca MC. Integrative Analysis of Cytokine and Lipidomics Datasets Following Mild Traumatic Brain Injury in Rats. Metabolites. 2024; 14(3):133. https://doi.org/10.3390/metabo14030133
Chicago/Turabian StylePulliam, Alexis N., Alyssa F. Pybus, David A. Gaul, Samuel G. Moore, Levi B. Wood, Facundo M. Fernández, and Michelle C. LaPlaca. 2024. "Integrative Analysis of Cytokine and Lipidomics Datasets Following Mild Traumatic Brain Injury in Rats" Metabolites 14, no. 3: 133. https://doi.org/10.3390/metabo14030133