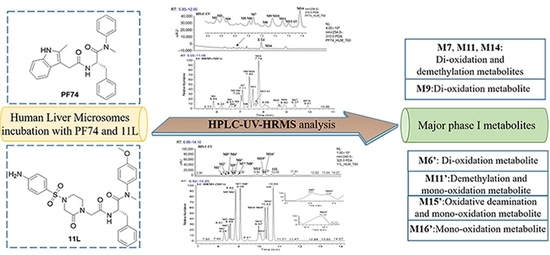
Metabolite Identification of HIV-1 Capsid Modulators PF74 and 11L in Human Liver Microsomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Preparation
2.3. Chromatographic and Mass Spectrometric Methods
2.3.1. Chromatographic and Mass Spectrometric Methods of PF74
2.3.2. Chromatographic and Mass Spectrometric Methods of 11L
2.4. Data Processing
3. Results and Discussion
3.1. Identification of PF74 Metabolites and Proposed Metabolic Pathways in HLMs
3.1.1. Metabolite Profiling of PF74 in HLMs
Amide Hydrolysis Metabolite (M1)
Tri-oxidation and Demethylation Metabolites (M2, M3, M6, M8, M10, and M12)
Tri-oxidation Metabolites (M4 and M5)
Di-oxidation and Demethylation Metabolites (M7, M11, M13, M14, and M16)
Di-oxidation Metabolite (M9)
Mono-oxidation Metabolite (M15)
Mono-oxidation and Demethylation Metabolite (M17)
3.1.2. Proposed Metabolic Pathways of PF74
3.2. Identification of 11L Metabolites and Proposed Metabolic Pathways in HLMs
3.2.1. Metabolite Profiling of 11L in HLMs
Tri-oxidation Metabolite (M1′)
Di-oxidation Metabolites (M2′, M6′, M13′)
Demethylation and Mono-oxidation Metabolites (M3′, M9′, M10′, M11′)
Demethylation and Di-oxidation Metabolite (M4′)
N-demethylation and O-demethylation Metabolite (M5′)
Mono-oxidation Metabolites (M7′, M16′)
N-demethylation Metabolite (M8′)
Dehydrogenation and Mono-oxidation Metabolite (M12′)
Demethylation Metabolite (M14′)
Oxidative Deamination and Mono-oxidation Metabolite (M15′)
3.2.2. Proposed Metabolic Pathways of 11L
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davenport, M.P.; Khoury, D.S.; Cromer, D.; Lewin, S.R.; Kelleher, A.D.; Kent, S.J. Functional cure of HIV: The scale of the challenge. Nat. Rev. Immunol. 2019, 19, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Kleinpeter, A.B.; Freed, E.O. HIV-1 Maturation: Lessons Learned from Inhibitors. Viruses 2020, 12, 940. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Pannecouque, C.; De Clercq, E.; Liu, X. Anti-HIV Drug Discovery and Development: Current Innovations and Future Trends. J. Med. Chem. 2016, 59, 2849–2878. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Das, D.; Kobayakawa, T.; Tamamura, H.; Takeuchi, H. Discovery and Development of Anti-HIV Therapeutic Agents: Progress Towards Improved HIV Medication. Curr. Top. Med. Chem. 2019, 19, 1621–1649. [Google Scholar] [CrossRef]
- Xu, S.; Sun, L.; Huang, B.; Liu, X.; Zhan, P. Medicinal chemistry strategies of targeting HIV-1 capsid protein for antiviral treatment. Future Med. Chem. 2020, 12, 1281–1284. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, S.; Sun, L.; Ding, D.; Tao, Y.; Kang, D.; Liu, X.; Zhan, P. HIV-1 capsid inhibitors: A sword to destroy the virus. Future Med. Chem. 2022, 14, 605–607. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.; Xu, S.; Huang, T.; Song, S.; Cherukupalli, S.; Zhan, P.; Liu, X. An insight on medicinal aspects of novel HIV-1 capsid protein inhibitors. Eur. J. Med. Chem. 2021, 217, 113380. [Google Scholar] [CrossRef]
- Shi, J.; Zhou, J.; Shah, V.B.; Aiken, C.; Whitby, K. Small-Molecule Inhibition of Human Immunodeficiency Virus Type 1 Infection by Virus Capsid Destabilization. J. Virol. 2011, 85, 542–549. [Google Scholar] [CrossRef]
- Balasubramaniam, M.; Zhou, J.; Addai, A.; Martinez, P.; Pandhare, J.; Aiken, C.; Dash, C. PF74 Inhibits HIV-1 Integration by Altering the Composition of the Preintegration Complex. J. Virol. 2019, 93, e01741-18. [Google Scholar] [CrossRef]
- Sun, L.; Dick, A.; Meuser, M.E.; Huang, T.; Zalloum, W.A.; Chen, C.H.; Cherukupalli, S.; Xu, S.; Ding, X.; Gao, P.; et al. Design, Synthesis, and Mechanism Study of Benzenesulfonamide-Containing Phenylalanine Derivatives as Novel HIV-1 Capsid Inhibitors with Improved Antiviral Activities. J. Med. Chem. 2020, 63, 4790–4810. [Google Scholar] [CrossRef]
- Pak, A.J.; Grime, J.M.A.; Yu, A.; Voth, G.A. Off-Pathway Assembly: A Broad-Spectrum Mechanism of Action for Drugs That Undermine Controlled HIV-1 Viral Capsid Formation. J. Am. Chem. Soc. 2019, 141, 10214–10224. [Google Scholar] [CrossRef] [PubMed]
- Baranczewski, P.; Stańczak, A.; Kautiainen, A.; Sandin, P.; Edlund, P.-O. Introduction to early in vitro identification of metabolites of new chemical entities in drug discovery and development. Pharmacol. Rep. 2006, 58, 341. [Google Scholar] [PubMed]
- Słoczyńska, K.; Gunia-Krzyżak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Żelaszczyk, D.; Popiół, J.; Pękala, E. Metabolic stability and its role in the discovery of new chemical entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Trunzer, M.; Faller, B.; Zimmerlin, A. Metabolic Soft Spot Identification and Compound Optimization in Early Discovery Phases Using MetaSite and LC-MS/MS Validation. J. Med. Chem. 2009, 52, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Padilha, E.C.; Shah, P.; Rai, G.; Xu, X. NOX2 inhibitor GSK2795039 metabolite identification towards drug optimization. J. Pharm. Biomed. Anal. 2021, 201, 114102. [Google Scholar] [CrossRef]
- Lee, K.; Lee, J.-Y.; Lee, K.; Jung, C.-R.; Kim, M.J.; Kim, J.A.; Yoo, D.G.; Shin, E.J.; Oh, S.J. Metabolite Profiling and Characterization of LW6, a Novel HIF-1α Inhibitor, as an Antitumor Drug Candidate in Mice. Molecules 2021, 26, 1951. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Ndjoko, K.; Hostettmann, K. Liquid chromatography with ultraviolet absorbance-mass spectrometric detection and with nuclear magnetic resonance spectroscopy: A powerful combination for the on-line structural investigation of plant metabolites. J. Chromatogr. A 2003, 1000, 437–455. [Google Scholar] [CrossRef]
- Chen, Y.; Monshouwer, M.; Fitch, W.L. Analytical Tools and Approaches for Metabolite Identification in Early Drug Discovery. Pharm. Res. 2007, 24, 248–257. [Google Scholar] [CrossRef]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta. Med. 2016, 82, 857–871. [Google Scholar]
- Breindahl, T.; Evans-Brown, M.; Hindersson, P.; McVeigh, J.; Bellis, M.; Stensballe, A.; Kimergård, A. Identification and characterization by LC-UV-MS/MS of melanotan II skin-tanning products sold illegally on the Internet. Drug Test. Anal. 2015, 7, 164–172. [Google Scholar] [CrossRef]
- Liang, Y.; Zhao, W.; Wang, C.; Wang, Z.; Wang, Z.; Zhang, J. A Comprehensive Screening and Identification of Genistin Metabolites in Rats Based on Multiple Metabolite Templates Combined with UHPLC-HRMS Analysis. Molecules 2018, 23, 1862. [Google Scholar] [CrossRef] [PubMed]
- Paudel, S.; Kim, Y.; Choi, S.M.; Kim, J.; Bae, J.; Lee, T.; Lee, S. Identification of suberosin metabolites in human liver microsomes by high-performance liquid chromatography combined with high-resolution quadrupole–orbitrap mass spectrometer. Biol. Mass Spectrom. 2021, 56, e4623. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, L.; Deng, P.; Chen, X.; Zhong, D. Characterization of metabolites of GLS4 in humans using ultrahigh-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 2483–2492. [Google Scholar] [CrossRef]
- Montesano, C.; Vincenti, F.; Fanti, F.; Marti, M.; Bilel, S.; Togna, A.; Gregori, A.; Di Rosa, F.; Sergi, M. Untargeted Metabolic Profiling of 4-Fluoro-Furanylfentanyl and Isobutyrylfentanyl in Mouse Hepatocytes and Urine by Means of LC-HRMS. Metabolites 2021, 11, 97. [Google Scholar] [CrossRef]
- Wang, L.; Casey, M.C.; Vernekar, S.K.V.; Sahani, R.L.; Kirby, K.A.; Du, H.; Zhang, H.; Tedbury, P.R.; Xie, J.; Sarafianos, S.G.; et al. Novel PF74-like small molecules targeting the HIV-1 capsid protein: Balance of potency and metabolic stability. Acta Pharm. Sin. B 2021, 11, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Sun, L.; Dick, A.; Zalloum, W.A.; Huang, T.; Meuser, M.E.; Zhang, X.; Tao, Y.; Cherukupalli, S.; Ding, D.; et al. Design, synthesis, and mechanistic investigations of phenylalanine derivatives containing a benzothiazole moiety as HIV-1 capsid inhibitors with improved metabolic stability. Eur. J. Med. Chem. 2022, 227, 113903. [Google Scholar] [CrossRef] [PubMed]
- Sahani, R.; Diana-Rivero, R.; Vernekar, S.; Wang, L.; Du, H.; Zhang, H.; Castaner, A.; Casey, M.; Kirby, K.; Tedbury, P.; et al. Design, Synthesis and Characterization of HIV-1 CA-Targeting Small Molecules: Conformational Restriction of PF74. Viruses 2021, 13, 479. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sánchez-Roselló, M.; Aceña, J.L.; del Pozo, C.; Sorochinsky, A.E.; Fustero, S.; Soloshonok, V.A.; Liu, H. Fluorine in pharmaceutical industry: Fluorine-containing drugs introduced to the market in the last decade (2001–2011). Chem. Rev. 2014, 114, 2432–2506. [Google Scholar] [CrossRef]
- Sahani, R.L.; Akther, T.; Cilento, M.E.; Castaner, A.E.; Zhang, H.; Kirby, K.A.; Xie, J.; Sarafianos, S.G.; Wang, Z. Potency and metabolic stability: A molecular hybrid case in the design of novel PF74-like small molecules targeting HIV-1 capsid protein. RSC Med. Chem. 2021, 12, 2031–2044. [Google Scholar] [CrossRef]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364. [Google Scholar] [CrossRef]
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 2020, 584, 614–618. [Google Scholar] [CrossRef] [PubMed]









| Metabolite | [M + H]+ m/z | Retention Time (min) | Relative Abundance (UV Peak Area%) | UV Peak Area (254–310 nm) | Type of Biotransformation | Diagnostic Ions |
|---|---|---|---|---|---|---|
| M1 | 255.1490 | 5.85 | + | + | Amide hydrolysis (P − C11H9NO) | 255.15, 120.08 |
| M2 | 460.1866 | 6.57 | 4.26 | 1269 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 442.18, 351.13, 323.14, 257.13, 204.07, 190.09, 186.05, 162.05, 120.08 |
| M3 | 460.1866 | 6.61 | 4.71 | 1401 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 442.18, 206.08, 204.07, 186.05, 162.05, 136.08 |
| M4 # | 474.2017 | 6.68 | 3.54 | 1055 | Tri-oxidation (P + 3O) | 474.20, 456.19, 222.08, 188.07, 170.06, 152.07, 108.08 |
| M5 * | 474.2028 | 7.05 | 4.97 | 1480 | Tri-oxidation (P + 3O) | 474.20, 456.19, 202.05, 174.06, 120.08, 108.08 |
| M6 * | 460.1866 | 7.05 | 2.39 | 713 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 442.18, 367.13, 339.13, 222.08, 188.07, 170.06, 152.07 |
| M7 | 444.1917 | 7.11 | 13.73 | 4088 | Di-oxidation and demethylation (P + 2O − CH2) | 444.19, 257.13, 188.07, 170.06, 120.08 |
| M8 | 460.1867 | 7.29 | 2.67 | 796 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 442.18, 202.05, 186.05 |
| M9 * | 458.2074 | 7.40 | 13.48 | 4013 | Di-oxidation(P + 2O) | 458.21, 440.20, 333.12, 305.13, 204.07, 190.09, 162.05, 108.08 |
| M10 * | 460.1863 | 7.40 | 2.87 | 854 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 442.20, 351.13, 323.14, 257.13, 204.07, 190.09, 186.05, 162.05, 120.08 |
| M11 | 444.1917 | 7.56 | 9.81 | 2919 | Di-oxidation and demethylation (P + 2O − CH2) | 444.19, 204.07, 186.06, 162.05 |
| M12 * | 460.1864 | 7.65 | 1.35 | 403 | Tri-oxidation and demethylation (P + 3O − CH2) | 460.19, 367.13, 339.13 |
| M13 * | 444.1918 | 7.65 | 4.94 | 1471 | Di-oxidation and demethylation (P + 2O − CH2) | 444.19, 257.13, 188.07, 170.06, 120.08 |
| M14 | 444.1915 | 7.79 | 25.62 | 7627 | Di-oxidation and demethylation (P + 2O − CH2) | 444.19, 204.07, 186.06, 162.05 |
| M15 | 442.2134 | 8.75 | + | + | Mono-oxidation (P + O) | 442.21, 424.20, 317.13, 289.13, 170.06 |
| M16 | 444.1919 | 8.90 | 4.29 | 1279 | Di-oxidation and demethylation (P + 2O − CH2) | 444.19, 204.07, 186.06, 162.06 |
| M17 | 428.1969 | 9.37 | 1.34 | 399 | Mono-oxidation and demethylation (P + O − CH2) | 428.20, 410.19, 317.18, 289.13, 188.07, 170.06 |
| PF74 (Parent) | 426.2169 | 9.91 | + | + | NA | 426.22, 319.14, 291.15, 172.08, 144.08, 120.08 |
| Metabolite | Formula | Exact Mass | Measured Value (m/z) | Theoretical Value (m/z) | PPM |
|---|---|---|---|---|---|
| M1 | C16H18N2O | 254.1419 | 255.1490 | 255.1492 | −0.8 |
| M2 | C26H25N3O5 | 459.1794 | 460.1866 | 460.1867 | −0.2 |
| M3 | C26H25N3O5 | 459.1794 | 460.1866 | 460.1867 | −0.2 |
| M4 | C27H27N3O5 | 473.1951 | 474.2017 | 474.2024 | −1.5 |
| M5 | C27H27N3O5 | 473.1951 | 474.2028 | 474.2024 | 0.8 |
| M6 | C26H25N3O5 | 459.1794 | 460.1866 | 460.1867 | −0.2 |
| M7 | C26H25N3O4 | 443.1845 | 444.1917 | 444.1918 | −0.2 |
| M8 | C26H25N3O5 | 459.1794 | 460.1867 | 460.1867 | 0.0 |
| M9 | C27H27N3O4 | 457.2002 | 458.2074 | 458.2075 | −0.2 |
| M10 | C26H25N3O5 | 459.1794 | 460.1863 | 460.1867 | −0.9 |
| M11 | C26H25N3O4 | 443.1845 | 444.1917 | 444.1918 | −0.2 |
| M12 | C26H25N3O5 | 459.1794 | 460.1864 | 460.1867 | −0.7 |
| M13 | C26H25N3O4 | 443.1845 | 444.1918 | 444.1918 | 0.0 |
| M14 | C26H25N3O4 | 443.1845 | 444.1915 | 444.1918 | −0.7 |
| M15 | C27H27N3O3 | 441.2052 | 442.2134 | 442.2125 | 2.0 |
| M16 | C26H25N3O4 | 443.1845 | 444.1919 | 444.1918 | 0.2 |
| M17 | C26H25N3O3 | 427.1896 | 428.1969 | 428.1969 | 0.0 |
| PF74 | C27H27N3O2 | 425.2103 | 426.2169 | 426.2176 | −1.6 |
| Metabolite | [M + H]+/[M − H]− m/z | Retention Time (min) | Relative Abundance (UV Peak Area%) | UV Peak Area (240–320 nm) | Type of Biotransformation | Diagnostic Ions |
|---|---|---|---|---|---|---|
| M1′ | 628.2078 | 8.29 | 5.36 | 7217 | Tri-oxidation (P + 3O) | 628.21, 284.07, 138.09 |
| M2′ | 612.2122 | 8.49 | 5.36 | 7223 | Di-oxidation (P + 2O) | 612.21, 475.13, 296.07, 268.08, 138.09 |
| M3′ | 582.2014 | 8.56 | 6.81 | 9171 @ | Mono-oxidation and demethylation (P + O– CH2) | 582.20, 443.14, 296.07, 268.08, 140.07, |
| M4′ | 598.1962 | 8.60 | 0.77 | 1043 @ | Di-oxidation and demethylation (P+ 2O − CH2) | 598.20, 475.13, 312.07, 172.01, 122.06 |
| M5′ | 552.1917 | 8.79 | 4.94 | 6649 | N- demethylation and O- demethylation (P − 2CH2) | 552.19, 443.14, 296.07, 268.08 |
| M6′ | 612.2105 | 8.82 | 12.01 | 16,172 | Di-oxidation (P + 2O) | 612.21, 475.13, 312.07, 172.01 |
| M7′ | 596.2166 | 9.05 | 6.62 | 8923 @ | Mono-oxidation (P + O) | 596.22, 459.13, 296.07, 268.08, 138.09 |
| M8′ | 566.2041 | 9.08 | 2.25 | 3028 @ | N- demethylation (P − CH2) | 566.20, 124.08 |
| M9′ | 580.1873 * | 9.78 | 2.59 | 3493 | Mono-oxidation and demethylation (P + O– CH2) | 580.19, 156.01 |
| M10′ | 580.1874 * | 9.83 | 2.62 | 3533 | Mono-oxidation and demethylation (P + O– CH2) | 580.19, 156.01 |
| M11′ | 582.2020 | 9.98 | 8.77 | 11,814 | Mono-oxidation and demethylation (P + O − CH2) | 582.20, 172.01, 122.06 |
| M12′ | 594.2021 | 10.04 | 2.79 | 3755 | Mono-oxidation and dehydrogenation (P + O − 2H) | 594.20, 254.06, 172.01 |
| M13′ | 610.1976 * | 10.06 | 3.27 | 4410 | Di-oxidation (P + 2O) | 610.20, 270.06, 172.01 |
| M14′ | 566.2068 | 10.29 | 3.55 | 4777 # @ | Demethylation (P − CH2) | 566.20, 443.14, 296.07, 268.08, 156.01 |
| M15′ | 597.2002 | 10.32 | 15.98 | 21,520 # @ | Mono-oxidation and oxidative deamination(P + O − NH + O) | 597.20, 579.21, 313.05, 285.05 |
| M16′ | 596.2158 | 10.36 | 9.20 | 12,391 # @ | Mono-oxidation (P + O) | 596.22, 312.07, 284.07 |
| 11L (Parent) | 580.2224/578.2077 | 10.69 | 7.11 | 9581 | NA | 580.22, 443.14, 415.14, 296.07, 268.08, 240.08 (+); 578.21, 441.12, 311.08, 294.06, 254.06, 228.04, 156.01, 92.05 (−) |
| Metabolite | Formula | Exact Mass | Measured Value | Theoretical Value | PPM |
|---|---|---|---|---|---|
| (m/z) | (m/z) | ||||
| M1′ | C29H33N5O9S | 627.1999 | 628.2078 | 628.2072 | 1.0 |
| M2′ | C29H33N5O8S | 611.2050 | 612.2122 | 612.2123 | −0.2 |
| M3′ | C28H31N5O7S | 581.1944 | 582.2014 | 582.2017 | −0.5 |
| M4′ | C28H31N5O8S | 597.1893 | 598.1962 | 598.1966 | −0.7 |
| M5′ | C27H29N5O6S | 551.1839 | 552.1917 | 552.1912 | 0.9 |
| M6′ | C29H33N5O8S | 611.2050 | 612.2105 | 612.2123 | −2.9 |
| M7′ | C29H33N5O7S | 595.2101 | 596.2166 | 596.2174 | −1.3 |
| M8′ | C28H31N5O6S | 565.1995 | 566.2041 | 566.2068 | −4.8 |
| M9′ | C28H31N5O7S | 581.1944 | 580.1873 * | 580.1871 | 0.3 |
| M10′ | C28H31N5O7S | 581.1944 | 580.1874 * | 580.1871 | 0.5 |
| M11′ | C28H31N5O7S | 581.1944 | 582.2020 | 582.2017 | 0.5 |
| M12′ | C29H31N5O7S | 593.1944 | 594.2021 | 594.2017 | 0.7 |
| M13′ | C29H33N5O8S | 611.2050 | 610.1976 * | 610.1977 | −0.2 |
| M14′ | C28H31N5O6S | 565.1995 | 566.2068 | 566.2068 | 0.0 |
| M15′ | C29H32N4O8S | 596.1941 | 597.2002 | 597.2014 | −2.0 |
| M16′ | C29H33N5O7S | 595.2101 | 596.2158 | 596.2174 | −2.7 |
| 11L | C29H33N5O6S | 579.2152 | 580.2224 | 580.2225 | −0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Sun, L.; Ding, D.; Zhang, X.; Liu, X.; Zhan, P. Metabolite Identification of HIV-1 Capsid Modulators PF74 and 11L in Human Liver Microsomes. Metabolites 2022, 12, 752. https://doi.org/10.3390/metabo12080752
Xu S, Sun L, Ding D, Zhang X, Liu X, Zhan P. Metabolite Identification of HIV-1 Capsid Modulators PF74 and 11L in Human Liver Microsomes. Metabolites. 2022; 12(8):752. https://doi.org/10.3390/metabo12080752
Chicago/Turabian StyleXu, Shujing, Lin Sun, Dang Ding, Xujie Zhang, Xinyong Liu, and Peng Zhan. 2022. "Metabolite Identification of HIV-1 Capsid Modulators PF74 and 11L in Human Liver Microsomes" Metabolites 12, no. 8: 752. https://doi.org/10.3390/metabo12080752