Exercise and Interorgan Communication: Short-Term Exercise Training Blunts Differences in Consecutive Daily Urine 1H-NMR Metabolomic Signatures between Physically Active and Inactive Individuals
Abstract
:1. Introduction
2. Results and Discussion
2.1. Integrated Analysis of Exercise Data and the X-Adapt Urine-Sample Collection
2.2. Differences in Urine 1H-NMR Metabolomes between the Trained and Untrained Groups: The Extended Urine-Sample Collection
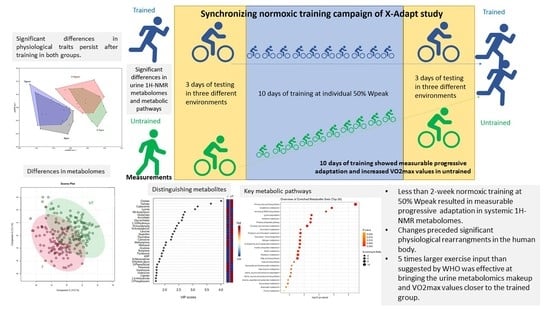
2.3. Differences between Trained and Untrained Groups before and after Synchronizing Normoxic Training Campaign: The Extended Urine-Sample Collection
3. Materials and Methods
3.1. Project Description
3.2. Sample Collection
3.3. NMR Metabolomics
3.4. Physicochemical Parameters of Urine Samples
3.5. Statistical Analysis and Machine Learning
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelly, R.S.; Kelly, M.P.; Kelly, P. Metabolomics, physical activity, exercise and health: A review of the current evidence. Biochim. Biophys. Acta. Mol. Basis Dis. 2020, 1866, 165936. [Google Scholar] [CrossRef]
- Sallis, J.F.; Bull, F.; Guthold, R.; Heath, G.W.; Inoue, S.; Kelly, P.; Oyeyemi, A.L.; Perez, L.G.; Richards, J.; Hallal, P.C.; et al. Progress in physical activity over the Olympic quadrennium. Lancet 2016, 388, 1325–1336. [Google Scholar] [CrossRef]
- Bongiovanni, T.; Genovesi, F.; Nemmer, M.; Carling, C.; Alberti, G.; Howatson, G. Nutritional interventions for reducing the signs and symptoms of exercise-induced muscle damage and accelerate recovery in athletes: Current knowledge, practical application and future perspectives. Eur. J. Appl. Physiol. 2020, 120, 1965–1996. [Google Scholar] [CrossRef]
- Sket, R.; Debevec, T.; Kublik, S.; Schloter, M.; Schoeller, A.; Murovec, B.; Mikus, K.V.; Makuc, D.; Pecnik, K.; Plavec, J.; et al. Intestinal Metagenomes and Metabolomes in Healthy Young Males: Inactivity and Hypoxia Generated Negative Physiological Symptoms Precede Microbial Dysbiosis. Front. Physiol. 2018, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Šket, R.; Deutsch, L.; Prevoršek, Z.; Mekjavić, I.B.; Plavec, J.; Rittweger, J.; Debevec, T.; Eiken, O.; Stres, B. Systems View of Deconditioning During Spaceflight Simulation in the PlanHab Project: The Departure of Urine 1 H-NMR Metabolomes From Healthy State in Young Males Subjected to Bedrest Inactivity and Hypoxia. Front. Physiol. 2020, 11, 532271. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Asayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med.-Open 2018, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Danaher, J.; Gerber, T.; Wellard, R.M.; Stathis, C.G.; Cooke, M.B. The use of metabolomics to monitor simultaneous changes in metabolic variables following supramaximal low volume high intensity exercise. Metabolomics 2015, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Zafeiridis, A.; Chatziioannou, A.C.; Sarivasiliou, H.; Kyparos, A.; Nikolaidis, M.G.; Vrabas, I.S.; Pechlivanis, A.; Zoumpoulakis, P.; Baskakis, C.; Dipla, K.; et al. Global Metabolic Stress of Isoeffort Continuous and High Intensity Interval Aerobic Exercise: A Comparative 1 H NMR Metabonomic Study. J. Proteome Res. 2016, 15, 4452–4463. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Q.; Moore, S.C.; Keadle, S.K.; Xiang, Y.B.; Zheng, W.; Peters, T.M.; Leitzmann, M.F.; Ji, B.T.; Sampson, J.N.; Shu, X.O.; et al. Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. Int. J. Epidemiol. 2016, 45, 1433–1444. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Murovec, B.; Makuc, D.; Kolbl Repinc, S.; Prevorsek, Z.; Zavec, D.; Sket, R.; Pecnik, K.; Plavec, J.; Stres, B. 1H NMR metabolomics of microbial metabolites in the four MW agricultural biogas plant reactors: A case study of inhibition mirroring the acute rumen acidosis symptoms. J. Environ. Manag. 2018, 222, 428–435. [Google Scholar] [CrossRef]
- Sotiridis, A.; Debevec, T.; Ciuha, U.; McDonnell, A.C.; Mlinar, T.; Royal, J.T.; Mekjavic, I.B. Aerobic but not thermoregulatory gains following a 10-day moderate-intensity training protocol are fitness level dependent: A cross-adaptation perspective. Physiol. Rep. 2020, 8, e14355. [Google Scholar] [CrossRef] [Green Version]
- Jay, O.; Bain, A.R.; Deren, T.M.; Sacheli, M.; Cramer, M.N. Large differences in peak oxygen uptake do not independently alter changes in core temperature and sweating during exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R832–R841. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.; Lundby, C. Refuting the myth of non-response to exercise training: ‘non-responders’ do respond to higher dose of training. J. Physiol. 2017, 595, 3377–3387. [Google Scholar] [CrossRef] [PubMed]
- Sotiridis, A.; Debevec, T.; McDonnell, A.C.; Ciuha, U.; Eiken, O.; Mekjavic, I.B. Exercise cardiorespiratory and thermoregulatory responses in normoxic, hypoxic and hot environment following 10-day continuous hypoxic exposure. J. Appl. Physiol. 2018, 125, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Sotiridis, A.; Debevec, T.; Ciuha, U.; Eiken, O.; Mekjavic, I.B. Heat acclimation does not affect maximal aerobic power in thermoneutral normoxic or hypoxic conditions. Exp. Physiol. 2019, 104, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Sotiridis, A. Independent and Combined Effects of Heat and Hypoxic Acclimation on Exercise Performance in Humans: With Particular Reference to Cross-Adaption. Ph.D. Thesis, Jozef Stefan Institute, Ljubljana, Slovenia, 2019. [Google Scholar]
- Armstrong, N.; Barker, A.R. Endurance training and elite young athletes. Med. Sport Sci. 2011, 56, 59–83. [Google Scholar] [CrossRef]
- Siopi, A.; Deda, O.; Manou, V.; Kellis, S.; Kosmidis, I.; Komninou, D.; Raikos, N.; Christoulas, K.; Theodoridis, G.A.; Mougios, V. Effects of Different Exercise Modes on the Urinary Metabolic Fingerprint of Men with and without Metabolic Syndrome. Metabolites 2017, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Tsamardinos, I.; Charonyktakis, P.; Lakiotaki, K.; Borboudakis, G.; Zenklusen, J.C.; Juhl, H.; Chatzaki, E.; Lagani, V. Just Add Data: Automated Predictive Modeling and BioSignature Discovery. bioRxiv 2020, 1–46. [Google Scholar] [CrossRef]
- Spiering, B.A.; Kraemer, W.J.; Hatfield, D.L.; Vingren, J.L.; Fragala, M.S.; Ho, J.Y.; Thomas, G.A.; Häkkinen, K.; Volek, J.S. Effects of L-carnitine L-tartrate supplementation on muscle oxygenation responses to resistance exercise. J. Strength Cond. Res. 2008, 22, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Abramowicz, W.N.; Galloway, S.D. Effects of acute versus chronic L-carnitine L-tartrate supplementation on metabolic responses to steady state exercise in males and females. Int. J. Sport Nutr. Exerc. Metab. 2005, 15, 386–400. [Google Scholar] [CrossRef]
- Vázquez-Fresno, R.; Llorach, R.; Urpi-Sarda, M.; Khymenets, O.; Bulló, M.; Corella, D.; Fitó, M.; Martínez-González, M.A.; Estruch, R.; Andres-Lacueva, C. An NMR metabolomics approach reveals a combined-biomarkers model in a wine interventional trial with validation in free-living individuals of the PREDIMED study. Metabolomics 2014, 11, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Perez, I.; Posma, J.M.; Chambers, E.S.; Nicholson, J.K.; Mathers, J.C.; Beckmann, M.; Draper, J.; Holmes, E.; Frost, G. An Analytical Pipeline for Quantitative Characterization of Dietary Intake: Application To Assess Grape Intake. J. Agric. Food Chem. 2016, 64, 2423–2431. [Google Scholar] [CrossRef] [Green Version]
- Domínguez-López, I.; Parilli-Moser, I.; Arancibia-Riveros, C.; Tresserra-Rimbau, A.; Martínez-González, M.A.; Ortega-Azorín, C.; Salas-Salvadó, J.; Castañer, O.; Lapetra, J.; Arós, F.; et al. Urinary Tartaric Acid, a Biomarker of Wine Intake, Correlates with Lower Total and LDL Cholesterol. Nutrients 2021, 13, 2883. [Google Scholar] [CrossRef]
- Chen, J.J.; Xie, J.; Li, W.W.; Bai, S.J.; Wang, W.; Zheng, P.; Xie, P. Age-specific urinary metabolite signatures and functions in patients with major depressive disorder. Aging 2019, 11, 6626–6637. [Google Scholar] [CrossRef]
- Gerber, M.; Beck, J.; Brand, S.; Cody, R.; Donath, L.; Eckert, A.; Faude, O.; Fischer, X.; Hatzinger, M.; Holsboer-Trachsler, E.; et al. The impact of lifestyle Physical Activity Counselling in IN-PATients with major depressive disorders on physical activity, cardiorespiratory fitness, depression, and cardiovascular health risk markers: Study protocol for a randomized controlled trial. Trials 2019, 20, 367. [Google Scholar] [CrossRef] [Green Version]
- Debevec, T.; Bali, T.C.; Simpson, E.J.; Macdonald, I.A.; Eiken, O.; Mekjavic, I.B. Separate and combined effects of 21-day bed rest and hypoxic confinement on body composition. Eur. J. Appl. Physiol. 2014, 114, 2411–2425. [Google Scholar] [CrossRef]
- Debevec, T.; Simpson, E.J.; Mekjavic, I.B.; Eiken, O.; Macdonald, I.A. Effects of prolonged hypoxia and bed rest on appetite and appetite-related hormones. Appetite 2016, 107, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Sket, R.; Treichel, N.; Debevec, T.; Eiken, O.; Mekjavic, I.; Schloter, M.; Vital, M.; Chandler, J.; Tiedje, J.M.; Murovec, B.; et al. Hypoxia and Inactivity Related Physiological Changes (Constipation, Inflammation) Are Not Reflected at the Level of Gut Metabolites and Butyrate Producing Microbial Community: The PlanHab Study. Front. Physiol. 2017, 8, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sket, R.; Treichel, N.; Kublik, S.; Debevec, T.; Eiken, O.; Mekjavic, I.; Schloter, M.; Vital, M.; Chandler, J.; Tiedje, J.M.; et al. Hypoxia and inactivity related physiological changes precede or take place in absence of significant rearrangements in bacterial community structure: The PlanHab randomized trial pilot study. PLoS ONE 2017, 12, e0188556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debevec, T.; Ganse, B.; Mittag, U.; Eiken, O.; Mekjavic, I.B.; Rittweger, J. Hypoxia Aggravates Inactivity-Related Muscle Wasting. Front. Physiol. 2018, 9, 494. [Google Scholar] [CrossRef] [Green Version]
- Stavrou, N.A.M.; Debevec, T.; Eiken, O.; Mekjavic, I.B. Hypoxia Exacerbates Negative Emotional State during Inactivity: The Effect of 21 Days Hypoxic Bed Rest and Confinement. Front. Physiol. 2018, 9, 26. [Google Scholar] [CrossRef] [Green Version]
- Stavrou, N.A.M.; Debevec, T.; Eiken, O.; Mekjavic, I.B. Hypoxia Worsens Affective Responses and Feeling of Fatigue During Prolonged Bed Rest. Front. Psychol. 2018, 9, 362. [Google Scholar] [CrossRef]
- Strewe, C.; Zeller, R.; Feuerecker, M.; Hoerl, M.; Kumprej, I.; Crispin, A.; Johannes, B.; Debevec, T.; Mekjavic, I.; Schelling, G.; et al. PlanHab study: Assessment of psycho-neuroendocrine function in male subjects during 21 d of normobaric hypoxia and bed rest. Stress 2017, 20, 131–139. [Google Scholar] [CrossRef]
- Strewe, C.; Zeller, R.; Feuerecker, M.; Hoerl, M.; Matzel, S.; Kumprej, I.; Crispin, A.; Johannes, B.; Debevec, T.; Mekjavic, I.B.; et al. PlanHab Study: Consequences of combined normobaric hypoxia and bed rest on adenosine kinetics. Sci. Rep. 2018, 8, 1762. [Google Scholar] [CrossRef] [Green Version]
- Keramidas, M.E.; Kolegard, R.; Mekjavic, I.B.; Eiken, O. PlanHab: Hypoxia exaggerates the bed-rest-induced reduction in peak oxygen uptake during upright cycle ergometry. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H453–H464. [Google Scholar] [CrossRef]
- Louwies, T.; Jaki Mekjavic, P.; Cox, B.; Eiken, O.; Mekjavic, I.B.; Kounalakis, S.; De Boever, P. Separate and Combined Effects of Hypoxia and Horizontal Bed Rest on Retinal Blood Vessel Diameters. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4927–4932. [Google Scholar] [CrossRef] [Green Version]
- Simpson, E.J.; Debevec, T.; Eiken, O.; Mekjavic, I.; Macdonald, I.A. PlanHab: The combined and separate effects of 16 days of bed rest and normobaric hypoxic confinement on circulating lipids and indices of insulin sensitivity in healthy men. J. Appl. Physiol. 2016, 120, 947–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarabon, N.; Mekjavic, I.B.; Eiken, O.; Babic, J. The Effect of Bed Rest and Hypoxic Environment on Postural Balance and Trunk Automatic (Re)Actions in Young Healthy Males. Front. Physiol. 2018, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rullman, E.; Mekjavic, I.B.; Fischer, H.; Eiken, O. PlanHab (Planetary Habitat Simulation): The combined and separate effects of 21 days bed rest and hypoxic confinement on human skeletal muscle miRNA expression. Physiol. Rep. 2016, 4, e12753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrigo, J.; Gonzalez, F.; Aguirre, F.; Tacchi, F.; Gonzalez, A.; Meza, M.P.; Simon, F.; Cabrera, D.; Arrese, M.; Karpen, S.; et al. Cholic acid and deoxycholic acid induce skeletal muscle atrophy through a mechanism dependent on TGR5 receptor. J. Cell. Physiol. 2021, 236, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ung, T.T.; Nguyen, T.T.; Sah, D.K.; Park, S.Y.; Jung, Y.D. Cholic Acid Stimulates MMP-9 in Human Colon Cancer Cells via Activation of MAPK, AP-1, and NF-κB Activity. Int. J. Mol. Sci. 2020, 21, 3420. [Google Scholar] [CrossRef]
- Mercer, K.E.; Maurer, A.; Pack, L.M.; Ono-Moore, K.; Spray, B.J.; Campbell, C.; Chandler, C.J.; Burnett, D.; Souza, E.; Casazza, G.; et al. Exercise training and diet-induced weight loss increase markers of hepatic bile acid (BA) synthesis and reduce serum total BA concentrations in obese women. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E864–E873. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, T.; Zhao, A.; Ning, Z.; Kuang, J.; Wang, S.; You, Y.; Bao, Y.; Ma, X.; Yu, H.; et al. Hyocholic acid species as novel biomarkers for metabolic disorders. Nat. Commun. 2021, 12, 1487. [Google Scholar] [CrossRef]
- Alamoudi, J.A.; Li, W.; Gautam, N.; Olivera, M.; Meza, J.; Mukherjee, S.; Alnouti, Y. Bile acid indices as biomarkers for liver diseases I: Diagnostic markers. World J. Hepatol. 2021, 13, 433–455. [Google Scholar] [CrossRef]
- Pushpass, R.G.; Alzoufairi, S.; Jackson, K.G.; Lovegrove, J.A. Circulating bile acids as a link between the gut microbiota and cardiovascular health: Impact of prebiotics, probiotics and polyphenol-rich foods. Nutr. Res. Rev. 2021, 1–20. [Google Scholar] [CrossRef]
- Bathena, S.P.; Thakare, R.; Gautam, N.; Mukherjee, S.; Olivera, M.; Meza, J.; Alnouti, Y. Urinary bile acids as biomarkers for liver diseases II. Signature profiles in patients. Toxicol. Sci. Off. J. Soc. Toxicol. 2015, 143, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Lyu, J.; Li, H.; Yin, D.; Zhao, M.; Sun, Q.; Guo, M. Analysis of eight bile acids in urine of gastric cancer patients based on covalent organic framework enrichment coupled with liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2021, 1653, 462422. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Edakkanambeth Varayil, J.; Devanna, S.; Murad, M.H.; Iyer, P.G. Physical activity is associated with reduced risk of gastric cancer: A systematic review and meta-analysis. Cancer Prev. Res. 2014, 7, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abioye, A.I.; Odesanya, M.O.; Abioye, A.I.; Ibrahim, N.A. Physical activity and risk of gastric cancer: A meta-analysis of observational studies. Br. J. Sports Med. 2015, 49, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Salvagno, G.L.; Tarperi, C.; Negrini, D.; Montagnana, M.; Festa, L.; Sanchis-Gomar, F.; Schena, F.; Lippi, G. Middle-distance running acutely influences the concentration and composition of serum bile acids: Potential implications for cancer risk? Oncotarget 2017, 8, 52775–52782. [Google Scholar] [CrossRef] [Green Version]
- Maurer, A.; Ward, J.L.; Dean, K.; Billinger, S.A.; Lin, H.; Mercer, K.E.; Adams, S.H.; Thyfault, J.P. Divergence in aerobic capacity impacts bile acid metabolism in young women. J. Appl. Physiol. 2020, 129, 768–778. [Google Scholar] [CrossRef]
- Darenskaya, M.A.; Chugunova, E.V.; Kolesnikov, S.I.; Grebenkina, L.A.; Semenova, N.V.; Nikitina, O.A.; Kolesnikova, L.I. Content of Carbonyl Compounds and Parameters of Glutathione Metabolism in Men with Type 1 Diabetes Mellitus at Preclinical Stages of Diabetic Nephropathy. Bull. Exp. Biol. Med. 2021, 171, 592–595. [Google Scholar] [CrossRef]
- Liu, R.; Li, Q.; Ma, R.; Lin, X.; Xu, H.; Bi, K. Determination of polyamine metabolome in plasma and urine by ultrahigh performance liquid chromatography-tandem mass spectrometry method: Application to identify potential markers for human hepatic cancer. Anal. Chim. Acta 2013, 791, 36–45. [Google Scholar] [CrossRef]
- Maráková, K.; Piešťanský, J.; Zelinková, Z.; Mikuš, P. Simultaneous determination of twelve biogenic amines in human urine as potential biomarkers of inflammatory bowel diseases by capillary electrophoresis–tandem mass spectrometry. J. Pharm. Biomed. Anal. 2020, 186, 113294. [Google Scholar] [CrossRef]
- Venäläinen, M.K.; Roine, A.N.; Häkkinen, M.R.; Vepsäläinen, J.J.; Kumpulainen, P.S.; Kiviniemi, M.S.; Lehtimäki, T.; Oksala, N.K.; Rantanen, T.K. Altered Polyamine Profiles in Colorectal Cancer. Anticancer. Res. 2018, 38, 3601–3607. [Google Scholar] [CrossRef]
- Tian, Q.; Corkum, A.E.; Moaddel, R.; Ferrucci, L. Metabolomic profiles of being physically active and less sedentary: A critical review. Metab. Off. J. Metab. Soc. 2021, 17, 68. [Google Scholar] [CrossRef]
- Castro, A.; Duft, R.G.; Ferreira, M.L.V.; Andrade, A.L.L.; Gáspari, A.F.; Silva, L.M.; Oliveira-Nunes, S.G.; Cavaglieri, C.R.; Ghosh, S.; Bouchard, C.; et al. Association of skeletal muscle and serum metabolites with maximum power output gains in response to continuous endurance or high-intensity interval training programs: The TIMES study—A randomized controlled trial. PLoS ONE 2019, 14, e0212115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.M.; Dasari, S.; Konopka, A.R.; Johnson, M.L.; Manjunatha, S.; Esponda, R.R.; Carter, R.E.; Lanza, I.R.; Nair, K.S. Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Cell Metab. 2017, 25, 581–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabone, M.; Bressa, C.; García-Merino, J.A.; Moreno-Pérez, D.; Van, E.C.; Castelli, F.A.; Fenaille, F.; Larrosa, M. The effect of acute moderate-intensity exercise on the serum and fecal metabolomes and the gut microbiota of cross-country endurance athletes. Sci. Rep. 2021, 11, 3558. [Google Scholar] [CrossRef] [PubMed]
- Lustgarten, M.S.; Price, L.L.; Chalé, A.; Fielding, R.A. Metabolites related to gut bacterial metabolism, peroxisome proliferator-activated receptor-alpha activation, and insulin sensitivity are associated with physical function in functionally-limited older adults. Aging Cell 2014, 13, 918–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coen, P.M.; Jubrias, S.A.; Distefano, G.; Amati, F.; Mackey, D.C.; Glynn, N.W.; Manini, T.M.; Wohlgemuth, S.E.; Leeuwenburgh, C.; Cummings, S.R.; et al. Skeletal muscle mitochondrial energetics are associated with maximal aerobic capacity and walking speed in older adults. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2013, 68, 447–455. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Wei, M.; Hu, M.; Yue, K.; Bi, R.; Zhai, S.; Pi, Z.; Song, F.; Liu, Z. Pharmacodynamic and urinary metabolomics studies on the mechanism of Schisandra polysaccharide in the treatment of Alzheimer’s disease. Food Funct. 2019, 10, 432–447. [Google Scholar] [CrossRef]
- Gronek, P.; Balko, S.; Gronek, J.; Zajac, A.; Maszczyk, A.; Celka, R.; Doberska, A.; Czarny, W.; Podstawski, R.; Clark, C.C.T.; et al. Physical Activity and Alzheimer’s Disease: A Narrative Review. Aging Dis. 2019, 10, 1282–1292. [Google Scholar] [CrossRef] [Green Version]
- Maalouf, N.M.; Cameron, M.A.; Moe, O.W.; Adams-Huet, B.; Sakhaee, K. Low urine pH: A novel feature of the metabolic syndrome. Clin. J. Am. Soc. Nephrol. CJASN 2007, 2, 883–888. [Google Scholar] [CrossRef]
- Kraut, J.A.; Madias, N.E. Metabolic Acidosis of CKD: An Update. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2016, 67, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Otaki, Y.; Watanabe, T.; Takahashi, H.; Hasegawa, H.; Honda, S.; Funayama, A.; Netsu, S.; Ishino, M.; Arimoto, T.; Shishido, T.; et al. Acidic urine is associated with poor prognosis in patients with chronic heart failure. Heart Vessel. 2013, 28, 735–741. [Google Scholar] [CrossRef]
- Shimodaira, M.; Okaniwa, S.; Nakayama, T. Fasting Single-Spot Urine pH Is Associated with Metabolic Syndrome in the Japanese Population. Med. Princ. Pract. Int. J. Kuwait Univ. Health Sci. Cent. 2017, 26, 433–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, S.; Tsuji, H.; Ohmoto, Y.; Amakawa, K.; Hsieh, S.D.; Arase, Y.; Nakajima, H. High serum uric acid level and low urine pH as predictors of metabolic syndrome: A retrospective cohort study in a Japanese urban population. Metab. Clin. Exp. 2012, 61, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Kistner, S.; Rist, M.J.; Krüger, R.; Döring, M.; Schlechtweg, S.; Bub, A. High-Intensity Interval Training Decreases Resting Urinary Hypoxanthine Concentration in Young Active Men-A Metabolomic Approach. Metabolites 2019, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Di Liegro, C.M.; Schiera, G.; Proia, P.; Di Liegro, I. Physical Activity and Brain Health. Genes 2019, 10, 720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutsch, L.; Osredkar, D.; Plavec, J.; Stres, B. Spinal Muscular Atrophy after Nusinersen Therapy: Improved Physiology in Pediatric Patients with No Significant Change in Urine, Serum, and Liquor 1H-NMR Metabolomes in Comparison to an Age-Matched, Healthy Cohort. Metabolites 2021, 11, 206. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, L.; Stres, B. The Importance of Objective Stool Classification in Fecal 1H-NMR Metabolomics: Exponential Increase in Stool Crosslinking Is Mirrored in Systemic Inflammation and Associated to Fecal Acetate and Methionine. Metabolites 2021, 11, 172. [Google Scholar] [CrossRef]
- Madrid-Gambin, F.; Oller-Moreno, S.; Fernandez, L.; Bartova, S.; Giner, M.P.; Joyce, C.; Ferraro, F.; Montoliu, I.; Moco, S.; Marco, S. AlpsNMR: An R package for signal processing of fully untargeted NMR-based metabolomics. Bioinformatics 2020, 36, 2943–2945. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Emwas, A.H.; Saccenti, E.; Gao, X.; McKay, R.T.; Dos Santos, V.A.P.M.; Roy, R.; Wishart, D.S. Recommended strategies for spectral processing and post-processing of 1D 1 H-NMR data of biofluids with a particular focus on urine. Metab. Off. J. Metab. Soc. 2018, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Legendre, L.F.J. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 24, p. 1006. [Google Scholar]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deutsch, L.; Sotiridis, A.; Murovec, B.; Plavec, J.; Mekjavic, I.; Debevec, T.; Stres, B. Exercise and Interorgan Communication: Short-Term Exercise Training Blunts Differences in Consecutive Daily Urine 1H-NMR Metabolomic Signatures between Physically Active and Inactive Individuals. Metabolites 2022, 12, 473. https://doi.org/10.3390/metabo12060473
Deutsch L, Sotiridis A, Murovec B, Plavec J, Mekjavic I, Debevec T, Stres B. Exercise and Interorgan Communication: Short-Term Exercise Training Blunts Differences in Consecutive Daily Urine 1H-NMR Metabolomic Signatures between Physically Active and Inactive Individuals. Metabolites. 2022; 12(6):473. https://doi.org/10.3390/metabo12060473
Chicago/Turabian StyleDeutsch, Leon, Alexandros Sotiridis, Boštjan Murovec, Janez Plavec, Igor Mekjavic, Tadej Debevec, and Blaž Stres. 2022. "Exercise and Interorgan Communication: Short-Term Exercise Training Blunts Differences in Consecutive Daily Urine 1H-NMR Metabolomic Signatures between Physically Active and Inactive Individuals" Metabolites 12, no. 6: 473. https://doi.org/10.3390/metabo12060473