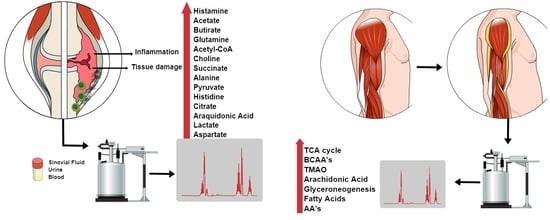
A Review of Metabolomic Profiling in Rheumatoid Arthritis: Bringing New Insights in Disease Pathogenesis, Treatment and Comorbidities
Abstract
:1. Introduction
2. Metabolomics
2.1. Techniques Used in Metabolomics
2.2. Metabolic Profile in Health and Disease
3. Metabolic Profile in RA
3.1. Lipidomic Profile in AR
3.2. Metabolites in the Synovial Fluid
| Sample | Sample Size (n) | Method Applied | Outcome | Reference |
|---|---|---|---|---|
| Synovial Fluid | 48 | GC/TOF MS | Positive correlation with DAS-28ES: radipate, fucose, glycocyamine, indole-3-lactate, isothreonate, phenylalanine and tryptophan asparagine Negative correlation with DAS-28ESR: citrate, cyano-/-alanine, oxoproline and ơ-alanine | [81] |
| Synovial Fluid | 38 | GC/TOF MS | Succinate, octadecanol, asparagine, terephthalate, salicylaldehyde, glutamine, citrulline, tyrosine, uracil, lysine, ribitol, tryptophan, xylose, ribose, isopalmitic acid, glycerol, myristic acid, palmitoleic acid, hydroxylamine and ethanolamine were validated as putative biomarkers for RA and discriminated from non-RA diseases | [84] |
| Synovial Fluid | 3 | LC-MS | Upregulated in RA: ibuprofen metabolism, glucocorticoid and mineralocorticoid metabolism, alpha-linolenic acid metabolism and steroid hormone biosynthesis. Downregulated in RA: purine and pyrimidine metabolism, arginine and proline metabolism; citrulline-nitric oxide cycle and glutathione metabolism. | [48,73] |
| Synovial Fluid | 20 | LC-MS | Activation of pyrimidine metabolism and purine metabolism, suppression of fatty acid biosynthesis and unsaturated fatty acid biosynthesis in RA | [105] |
| Blood | 25 | GC-MS | Decrease in histidine and threonic acid, methionine, asparagine, cholesterol in RA patients; Increase in glyceric acid, D-ribofuranose and hypoxanthine in RA patients | [82] |
| Plasma | 47 | 1H NMR spectroscopy | Cholesterol, lactate, acetylated glycoprotein, and lipid signatures were found to be possible biomarkers for disease severity | [85] |
| Plasma | 64 | UPLC-MS/MS | Acylcarnitine metabolites are increase in lower disease activity. Glucuronate and hypoxanthin were found to be significantly increased in higher disease activity | [95] |
| Plasma | 20 | GC-MS | L-cysteine, citric acid and L-glutamine | [92] |
| Serum | 53 | indirect calorimetry | Increases in metabolic rate in RA patients smokers compared to non-smokers patients | [83] |
| Serum | 27 | GC/TOF MS and UPLC−QTOF MS | Increases in homoserine, 4,8-dimethylnonanoyl carnitine, glyceraldehyde, lactic acid, dihydroxyfumaric acid and aspartic acid are shared between 4 types of arthritis | [87] |
| Serum | 58 | Spectrophotometer | RA patients presented methyl-histidine and hydroxyisocaproic acid, while hexose-phosphate and fructose-6-phosphate distinguished high ADA from low ADA | [94] |
| Serum | 124 | LC-MS/MS | Serum levels of NEFA (palmitic, stearic, palmitoleic, oleic, linoleic, γ-linoleic, AA, linolenic, EPA and docosahexaenoic–DHA). The NEFA profile in RA patients is associated with clinical characteristics of aggressive disease and enhanced Th1 response. | [88] |
| Serum | 33 | GC-MS | Disturbances of leucine, phenylalanine, pyroglutamate, serine, isoleucine, methionine, threonine, proline and valine), fatty acids (palmitelaidate, oleate, trans-9-octadecenoate, cis-5,8,11-eicosatrienoate, docosahexaenoate, 2-ketoisocaproate and 3-methyl-2-oxovalerate) and carbohydrates (mannose, ribose, scyllo-inositol, glycerol and 1,5-anhydrosorbitol) | [89] |
| Serum | 20 | 1H-NMR | Valine, isoleucine, lactate, alanine, creatinine, GPC APC and histidine relative levels were lower in RA, whereas 3-hydroxyisobutyrate, acetate, NAC, acetoacetate and acetone relative levels were higher compared with healthy controls. | [90] |
| Serum | 30 | LC-MS | 4-methoxyphenylacetic acid, glutamic acid, L-leucine, L-phenylalanine, L-tryptophan, L-proline, glyceraldehyde and fumaric acid are possible biomarkers for RA | [91] |
| Serum and urine | Serum (n = 126) and urine (n = 83) | NMR | Increased glycolysis, perturbation in the citrate cycle, oxidative stress, protein catabolism and increased urea cycle activity are present in newly presenting RA patients with elevated CRP. | [93] |
| Urine | 1400 | 1H-NMR | Lower levels of citrate were found in urine samples on RA patients | [86] |
3.3. Metabolites in Blood, Plasma and Serum of RA Patients
3.4. Metabolites in Urine Samples of RA Patients
3.5. Metabolites as Predictors of Disease Activity
4. Therapeutics of RA and Metabolic Profile
5. Metabolites and Comorbidities Associated with RA
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mcinnes, I.B.; Schett, G. The Pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, D.L.; Wolfe, F.; Huizinga, T.W.J. Rheumatoid Arthritis. Proc. Lancet 2010, 376, 1094–1108. [Google Scholar] [CrossRef]
- Feldmann, M.; Brennan, F.M. Rheumatoid Arthritis Review. Cell 1996, 85, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Choy, E. Understanding the dynamics: Pathways involved in the pathogenesis of rheumatoid arthritis. Rheumatology 2012, 51, v3–v11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerekes, G.; Nurmohamed, M.T.; González-Gay, M.A.; Seres, I.; Paragh, G.; Kardos, Z.; Baráth, Z.; Tamási, L.; Soltész, P.; Szekanecz, Z. Rheumatoid arthritis and metabolic syndrome. Nat. Rev. Rheumatol. 2014, 10, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.A.; Srikanth, V.K.; Fryer, J.L.; Blizzard, L.; Dwyer, T.; Venn, A.J. Dual energy X-ray absorptiometry body composition and aging in a population-based older cohort. Int. J. Obes. 2007, 31, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Santo, R.C.E.; Fernandes, K.Z.; Lora, P.S.; Filippin, L.I.; Xavier, R.M. Prevalence of rheumatoid cachexia in rheumatoid arthritis: A systematic review and meta-analysis. J. Cachexia. Sarcopenia Muscle 2018, 9, 816–825. [Google Scholar] [CrossRef] [Green Version]
- da Rocha, O.M.; Batista, A.d.A.P.; Maestá, N.; Burini, R.C.; Laurindo, I.M.M.; Kayser, C. Sarcopenia in Rheumatoid Cachexia: Definition, Mechanisms, Clinical Consequences and Potential Therapies. Rev. Bras. Reumatol. 2009, 49, 57–61. [Google Scholar]
- Walsmith, J.; Roubenoff, R. Cachexia in Rheumatoid Arthritis. Int. J. Cardiol. 2002, 85, 89–99. [Google Scholar] [CrossRef]
- Romão, V.C.; Fonseca, J.E. Etiology and Risk Factors for Rheumatoid Arthritis: A State-of-the-Art Review. Front. Med. 2021, 8, 689698. [Google Scholar] [CrossRef]
- Horning, E.C.; Horning, M.G. Metabolic Profiles: Gas-Phase Methods for Analysis of Metabolites. Clin. Chem. 1971, 17, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Alookaran, J.J.; Rhoads, J.M. Probiotics in Autoimmune and Inflammatory Disorders. Nutrients 2018, 10, 1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, D.G.; O’Neill, L.A. Krebs Cycle Reborn in Macrophage Immunometabolism. Annu. Rev. Immunol. 2020, 38, 289–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, J.K. Global systems biology, personalized medicine and molecular epidemiology. Mol. Syst. Biol. 2006, 2, 52. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Sadler, P.J.; Cain, K.; Holt, D.E.; Webbt, M.; Hawkes, G.E. 88 MHz I 3Cd-n.m.r. Studies of Native Rat Liver Metallothioneins. Biochem. J. 1983, 211, 251–255. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass Spectrometry Strategies in Metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S. Quantitative metabolomics using NMR. TrAC—Trends Anal. Chem. 2008, 27, 228–237. [Google Scholar] [CrossRef]
- Ravanbakhsh, S.; Liu, P.; Bjordahl, T.C.; Mandal, R.; Grant, J.R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; et al. Accurate, Fully-Automated NMR Spectral Profiling for Metabolomics. PLoS ONE 2015, 10, e0124219. [Google Scholar] [CrossRef] [Green Version]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2018, 37, 513–532. [Google Scholar] [CrossRef]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC—Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Selak, M.A.; Armour, S.M.; MacKenzie, E.D.; Boulahbel, H.; Watson, D.G.; Mansfield, K.D.; Pan, Y.; Simon, M.C.; Thompson, C.B.; Gottlieb, E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 2005, 7, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotstein, O.D.; Pruett, T.L.; Simmons, R.L. Mechanisms of Microbial Synergy in Polymicrobial Surgical Infections. Rev. Infect. Dis. 1985, 7, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ren, J.; Gao, D.S.; Dai, Y.; Yu, L. The Emerging Application of Itaconate: Promising Molecular Targets and Therapeutic Opportunities. Front. Chem. 2021, 9, 669308. [Google Scholar] [CrossRef] [PubMed]
- Bambouskova, M.; Gorvel, L.; Lampropoulou, V.; Sergushichev, A.; Loginicheva, E.; Johnson, K.; Korenfeld, D.; Mathyer, M.E.; Kim, H.; Huang, L.H.; et al. Electrophilic Properties of Itaconate and Derivatives Regulate the IkappaBzeta-ATF3 Inflammatory Axis (FAMIN) Is a Central Regulator of Immunometabolic Function. Nature 2018, 556, 201–504. [Google Scholar] [CrossRef] [PubMed]
- Cordes, T.; Lucas, A.; Divakaruni, A.S.; Murphy, A.N.; Cabrales, P.; Metallo, C.M. Itaconate modulates tricarboxylic acid and redox metabolism to mitigate reperfusion injury. Mol. Metab. 2020, 32, 122–135. [Google Scholar] [CrossRef]
- Lampropoulou, V.; Sergushichev, A.; Bambouskova, M.; Nair, S.; Vincent, E.E.; Loginicheva, E.; Cervantes-Barragan, L.; Ma, X.; Huang, S.C.C.; Griss, T.; et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016, 24, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Toma, I.; Kang, J.J.; Sipos, A.; Vargas, S.; Bansal, E.; Hanner, F.; Meer, E.; Peti-Peterdi, J. Succinate receptor GPR91 provides a direct link between high glucose levels and renin release in murine and rabbit kidney. J. Clin. Investig. 2008, 118, 2526–2534. [Google Scholar] [CrossRef] [Green Version]
- Sadagopan, N.; Li, W.; Roberds, S.L.; Major, T.; Preston, G.M.; Yu, Y.; Tones, M.A. Circulating Succinate Is Elevated in Rodent Models of Hypertension and Metabolic Disease. Am. J. Hypertens. 2007, 20, 1209–1215. [Google Scholar] [CrossRef]
- Velasquez, M.T.; Ramezani, A.; Manal, A.; Raj, D.S. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins 2016, 8, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Keogh, J.; Clifton, P. A review of potential metabolic etiologies of the observed association between red meat consumption and development of type 2 diabetes mellitus. Metabolism 2015, 64, 768–779. [Google Scholar] [CrossRef]
- Romano, K.A.; Vivas, E.I.; Amador-Noguez, D.; Rey, F.E. Intestinal Microbiota Composition Modulates Choline Bioavailability from Diet and Accumulation of the Proatherogenic Metabolite Trimethylamine-N-Oxide. MBio 2015, 6, e02481-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Dong, Y.; Ren, H.; Li, L.; He, C. A review of phytochemistry, metabolite changes, and medicinal uses of the common food mung bean and its sprouts (Vigna radiata). Chem. Cent. J. 2014, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, B.C.; Hullar, M.A.J.; Randolph, T.W.; Franke, A.A.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Shepherd, J.A.; Madeleine, M.M.; Le Marchand, L.; et al. Associations of plasma trimethylamine N-oxide, choline, carnitine, and betaine with inflammatory and cardiometabolic risk biomarkers and the fecal microbiome in the Multiethnic Cohort Adiposity Phenotype Study. Am. J. Clin. Nutr. 2020, 111, 1226–1234. [Google Scholar] [CrossRef]
- Macpherson, M.E.; Hov, J.R.; Ueland, T.; Dahl, T.B.; Kummen, M.; Otterdal, K.; Holm, K.; Berge, R.K.; Mollnes, T.E.; Trøseid, M.; et al. Gut Microbiota-Dependent Trimethylamine N-Oxide Associates With Inflammation in Common Variable Immunodeficiency. Front. Immunol. 2020, 11, 574500. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Brew, B.J.; Noonan, C.E.; Takikawa, O.; Cullen, K.M. Indoleamine 2,3 dioxygenase and quinolinic acid Immunoreactivity in Alzheimer’s disease hippocampus. Neuropathol. Appl. Neurobiol. 2005, 31, 395–404. [Google Scholar] [CrossRef]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sánchez, M.S.; Vázquez, C.; Peiró, C.; Egido, J.; Mas, S. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 Activation. Free Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Neishabouri, S.H.; Hutson, S.M.; Davoodi, J. Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino Acids 2015, 47, 1167–1182. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.; Makharia, G.; Ahuja, V.; Rajan, K.D.A.; Kalaivani, M.; Gupta, S.D.; Joshi, Y.K. Glutamine and Whey Protein Improve Intestinal Permeability and Morphology in Patients with Crohn’s Disease: A Randomized Controlled Trial. Dig. Dis. Sci. 2012, 57, 1000–1012. [Google Scholar] [CrossRef]
- Cruzat, V.; Rogero, M.M.; Keane, K.N.; Curi, R.; Newsholme, P. Glutamine: Metabolism and Immune Function, Supplementation and Clinical Translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wu, G.; Zhou, Z.; Dai, Z.; Sun, Y.; Ji, Y.; Li, W.; Wang, W.; Liu, C.; Han, F.; et al. Glutamine and intestinal barrier function. Amino Acids 2015, 47, 2143–2154. [Google Scholar] [CrossRef]
- Durante, W. The Emerging Role of L-Glutamine in Cardiovascular Health and Disease. Nutrients 2019, 11, 2092. [Google Scholar] [CrossRef] [Green Version]
- Eskildsen, M.P.; Hansen, P.B.L.; Stubbe, J.; Toft, A.; Walter, S.; Marcussen, N.; Rasmussen, L.M.; Vanhoutte, P.M.; Jensen, B.L. Prostaglandin I2 and Prostaglandin E2 Modulate Human Intrarenal Artery Contractility through Prostaglandin E2 -EP4, Prostacyclin-IP, and Thromboxane A2-TP Receptors. Hypertension 2014, 64, 551–556. [Google Scholar] [CrossRef] [Green Version]
- Gleim, S.; Stitham, J.; Tang, W.H.; Martin, K.A.; Hwa, J. An eicosanoid-centric view of atherothrombotic risk factors. Cell. Mol. Life Sci. 2012, 69, 3361–3380. [Google Scholar] [CrossRef] [Green Version]
- Shearer, G.C.; Newman, J.W. Impact of Circulating Esterifi Ed Eicosanoids and Other Oxylipins on Endothelial Function. Curr. Atheroscler. Rep. 2009, 11, 403–410. [Google Scholar] [CrossRef]
- Gio-Batta, M.; Sjöberg, F.; Jonsson, K.; Barman, M.; Lundell, A.C.; Adlerberth, I.; Hesselmar, B.; Sandberg, A.S.; Wold, A.E. Fecal short chain fatty acids in children living on farms and a link between valeric acid and protection from eczema. Sci. Rep. 2020, 10, 22449. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Golovko, S.; Golovko, M.Y.; Singh, S.; Darland, D.C.; Combs, C.K. Effects of Probiotic Supplementation on Short Chain Fatty Acids in the AppNL-G-F Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2020, 76, 1083–1102. [Google Scholar] [CrossRef] [PubMed]
- Valcheva, R.; Koleva, P.; Martínez, I.; Walter, J.; Gänzle, M.G.; Dieleman, L.A. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes 2019, 10, 334–357. [Google Scholar] [CrossRef] [PubMed]
- Ginos, B.N.R.; Navarro, S.L.; Schwarz, Y.; Gu, H.; Wang, D.; Randolph, T.W.; Shojaie, A.; Hullar, M.A.J.; Lampe, P.D.; Kratz, M.; et al. Circulating bile acids in healthy adults respond differently to a dietary pattern characterized by whole grains, legumes and fruits and vegetables compared to a diet high in refined grains and added sugars: A randomized, controlled, crossover feeding study. Metabolites 2018, 83, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Szántó, S.; Koreny, T.; Mikecz, K.; Glant, T.T.; Szekanecz, Z.; Varga, J. Inhibition of indoleamine 2,3-dioxygenase-mediated tryptophan catabolism accelerates collagen-induced arthritis in mice. Arthritis Res. Ther. 2007, 9, R50. [Google Scholar] [CrossRef] [Green Version]
- Mellor, A.L.; Lemos, H.; Huang, L. Indoleamine 2,3-Dioxygenase and Tolerance: Where Are We Now? Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Huang, I.-S.; Ogbechi, J.; Clanchy, F.I.; Williams, R.O.; Stone, T.W. IDO and Kynurenine Metabolites in Peripheral and CNS Disorders. Front. Immunol. 2020, 11, 388. [Google Scholar] [CrossRef] [Green Version]
- Sobczak, A.I.S.; Blindauer, C.A.; Stewart, A.J. Changes in Plasma Free Fatty Acids Associated with Type-2 Diabetes. Nutrients 2019, 11, 2022. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Carrio, J.; Salazar, N.; Margolles, A.; González, S.; Gueimonde, M.; de los Reyes-Gavilán, C.G.; Suárez, A. Free Fatty Acids Profiles Are Related to Gut Microbiota Signatures and Short-Chain Fatty Acids. Front. Immunol. 2017, 8, 823. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic Pathways in Immune Cell Activation and Quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Kummen, M.; Hov, J.R. The gut microbial influence on cholestatic liver disease. Liver Int. 2019, 39, 1186–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohmen, P.M.; Ozaki, S.; Verbeken, E.; Yperman, J.; Flameng, W.; Konertz, W.F. Tissue Engineering of an Auto-Xenograft Pulmonary Heart Valve. Asian Cardiovasc. Thorac. Ann. 2002, 10, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H. Immune Regulation by Microbiome Metabolites. Immunology 2018, 154, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Shanmuganathan, M.; Hart, L.; Pai, N.; Britz-Mckibbin, P. Urinary Metabolites Enable Differential Diagnosis and Therapeutic Monitoring of Pediatric Inflammatory Bowel Disease. Metabolites 2021, 11, 245. [Google Scholar] [CrossRef]
- Banfi, D.; Moro, E.; Bosi, A.; Bistoletti, M.; Cerantola, S.; Crema, F.; Maggi, F.; Giron, M.C.; Giaroni, C.; Baj, A. Impact of Microbial Metabolites on Microbiota–Gut–Brain Axis in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2021, 22, 1623. [Google Scholar] [CrossRef]
- Gorczyca, D.; Szponar, B.; Paściak, M.; Czajkowska, A.; Szmyrka, M. Serum levels of n-3 and n-6 polyunsaturated fatty acids in patients with systemic lupus erythematosus and their association with disease activity: A pilot study. Scand. J. Rheumatol. 2022, 51, 230–236. [Google Scholar] [CrossRef]
- Rosser, E.C.; Piper, C.J.M.; Matei, D.E.; Blair, P.A.; Rendeiro, A.F.; Orford, M.; Alber, D.G.; Krausgruber, T.; Catalan, D.; Klein, N.; et al. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab. 2020, 31, 837–851.e10. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; MacKay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Mizuno, M.; Noto, D.; Kaga, N.; Chiba, A.; Miyake, S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS ONE 2017, 12, e0173032. [Google Scholar] [CrossRef] [Green Version]
- Young, S.P.; Kapoor, S.R.; Viant, M.R.; Byrne, J.J.; Filer, A.; Buckley, C.D.; Kitas, G.D.; Raza, K. The Impact of Inflammation on Metabolomic Profiles in Patients with Arthritis. Arthritis Rheum. 2013, 65, 2015–2023. [Google Scholar] [CrossRef] [Green Version]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid arthritis. Nat. Rev. Dis. Prim. 2018, 4, 18001. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.D.; Filer, A. The role of the synovial fibroblast in rheumatoid arthritis pathogenesis. Curr. Opin. Rheumatol. 2015, 27, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Tolboom, T.C.A.; Van Der Helm-Van Mil, A.H.M.; Nelissen, R.G.H.H.; Breedveld, F.C.; Toes, R.E.M.; Huizinga, T.W.J. Invasiveness of fibroblast-like synoviocytes is an individual patient characteristic associated with the rate of joint destruction in patients with rheumatoid arthritis. Arthritis Rheum. 2005, 52, 1999–2002. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, M.; Balakrishnan, L.; Renuse, S.; Advani, J.; Goel, R.; Sathe, G.; Keshava Prasad, T.S.; Nair, B.; Jois, R.; Shankar, S.; et al. Synovial fluid proteome in rheumatoid arthritis. Clin. Proteomics 2016, 13, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, A.K.; Rawle, R.A.; Wallace, C.W.; Adams, E.; Greenwood, M.C.; Bothner, B.; June, R.K. Global metabolomic profiling of human synovial fluid for rheumatoid arthritis biomarkers. Clin. Exp. Rheumatol. 2019, 37, 393–399. [Google Scholar]
- Yang, W.; Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 2021, 18, 866–877. [Google Scholar] [CrossRef]
- Smolenska, Z.; Smolenski, R.T.; Zdrojewski, Z. Plasma concentrations of amino acid and nicotinamide metabolites in rheumatoid arthritis-potential biomarkers of disease activity and drug treatment. Biomarkers 2016, 21, 218–224. [Google Scholar] [CrossRef]
- Guma, M.; Tiziani, S.; Firestein, G.S. Metabolomics in rheumatic diseases: Desperately seeking biomarkers. Nat. Rev. Rheumatol. 2016, 12, 269–281. [Google Scholar] [CrossRef]
- Wymann, M.P.; Schneiter, R. Lipid Signalling in Disease. Nat. Rev. Mol. Cell Biol. 2008, 9, 162–176. [Google Scholar] [CrossRef]
- Chen, M.; Lam, B.K.; Kanaoka, Y.; Nigrovic, P.A.; Audoly, L.P.; Austen, K.F.; Lee, D.M. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J. Exp. Med. 2006, 203, 837–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polinski, K.J.; Bemis, E.A.; Yang, F.; Crume, T.; Demoruelle, M.K.; Feser, M.; Seifert, J.; O’Dell, J.R.; Mikuls, T.R.; Weisman, M.H.; et al. Association of Lipid Mediators With Development of Future Incident Inflammatory Arthritis in an Anti–Citrullinated Protein Antibody–Positive Population. Arthritis Rheumatol. Arthritis Rheumatol. 2021, 73, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Carrio, J.; Coras, R.; Alperi-López, M.; López, P.; Ulloa, C.; Ballina-García, F.J.; Armando, A.M.; Quehenberger, O.; Guma, M.; Suárez, A. Profiling of Serum Oxylipins During the Earliest Stages of Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.W.; Young, K.A.; Zerbe, G.O.; Kristen Demoruelle, M.; Weisman, M.H.; Buckner, J.H.; Gregersen, P.K.; Mikuls, T.R.; O’Dell, J.R.; Keating, R.M.; et al. Lower omega-3 fatty acids are associated with the presence of anti-cyclic citrullinated peptide autoantibodies in a population at risk for future rheumatoid arthritis: A nested case-control study. Rheumatology 2015, 55, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Chu, S.H.; Cui, J.; Sparks, J.A.; Lu, B.; Tedeschi, S.K.; Speyer, C.B.; Moss, L.K.; Feser, M.L.; Kelmenson, L.B.; Mewshaw, E.A.; et al. Circulating plasma metabolites and risk of rheumatoid arthritis in the Nurses’ Health Study. Rheumatology 2020, 59, 3369–3379. [Google Scholar] [CrossRef]
- Fuchs, B.; Schiller, J.; Wagner, U.; Häntzschel, H.; Arnold, K. The phosphatidylcholine/lysophosphatidylcholine ratio in human plasma is an indicator of the severity of rheumatoid arthritis: Investigations by 31P NMR and MALDI-TOF MS. Clin. Biochem. 2005, 38, 925–933. [Google Scholar] [CrossRef]
- Fuchs, B.; Bondzio, A.; Wagner, U.; Schiller, J. Phospholipid compositions of sera and synovial fluids from dog, human and horse: A comparison by 31p-Nmr and Maldi-Tof Ms. J. Anim. Physiol. Anim. Nutr. 2009, 93, 410–422. [Google Scholar] [CrossRef]
- Prete, P.E.; Gurakar-Osborne, A.; Kashyap, M.L. Synovial fluid lipids and apolipoproteins: A contemporary perspective. Biorheology 2017, 32, 1–16. [Google Scholar] [CrossRef]
- Hashimoto, A.; Hayashi, I.; Murakami, Y.; Sato, Y.; Kitasato, H.; Matsushita, R.; Iizuka, N.; Urabe, K.; Itoman, M.; Hirohata, S.; et al. Antiinflammatory mediator lipoxin A4 and its receptor in synovitis of patients with rheumatoid arthritis. J. Rheumatol. 2007, 34, 2144–2153. [Google Scholar]
- Rodgers, L.C.; Cole, J.; Rattigan, K.M.; Barrett, M.P.; Kurian, N.; McInnes, I.B.; Goodyear, C.S. The rheumatoid synovial environment alters fatty acid metabolism in human monocytes and enhances CCL20 secretion. Rheumatology 2019, 59, 869–878. [Google Scholar] [CrossRef]
- Giera, M.; Ioan-Facsinay, A.; Toes, R.; Gao, F.; Dalli, J.; Deelder, A.M.; Serhan, C.N.; Mayboroda, O.A. Lipid and lipid mediator profiling of human synovial fluid in rheumatoid arthritis patients by means of LC-MS/MS. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2012, 1821, 1415–1424. [Google Scholar] [CrossRef] [Green Version]
- Koh, J.H.; Yoon, S.J.; Kim, M.; Cho, S.; Lim, J.; Park, Y.; Kim, H.S.; Kwon, S.W.; Kim, W.U. Lipidome profile predictive of disease evolution and activity in rheumatoid arthritis. Exp. Mol. Med. 2022, 54, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Kiener, H.P.; Watts, G.F.M.; Cui, Y.; Wright, J.; Thornhill, T.S.; Sköld, M.; Behar, S.M.; Niederreiter, B.; Lu, J.; Cernadas, M.; et al. Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture. Arthritis Rheum. 2010, 62, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Filer, A. The fibroblast as a therapeutic target in rheumatoid arthritis. Curr. Opin. Pharmacol. 2013, 13, 413–419. [Google Scholar] [CrossRef]
- Bartok, B.; Firestein, G.S. Fibroblast-like synoviocytes: Key effector cells in rheumatoid arthritis. Immunol. Rev. 2010, 233, 233–255. [Google Scholar] [CrossRef] [PubMed]
- Bottini, N.; Firestein, G.S. Duality of fibroblast-like synoviocytes in RA: Passive responders and imprinted aggressors. Nat. Rev. Rheumatol. 2013, 9, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Misharin, A.V.; Cuda, C.M.; Saber, R.; Turner, J.D.; Gierut, A.K.; Kenneth Haines, G.K.; Berdnikovs, S.; Filer, A.; Clark, A.R.; Buckley, C.D.; et al. Nonclassical Ly6C-Monocytes Drive the Development of Inflammatory Arthritis in Mice. Cell Rep. 2014, 9, 591–604. [Google Scholar] [CrossRef] [Green Version]
- Shime, H.; Yabu, M.; Akazawa, T.; Kodama, K.; Matsumoto, M.; Seya, T.; Inoue, N. Tumor-Secreted Lactic Acid Promotes IL-23/IL-17 Proinflammatory Pathway. J. Immunol. 2008, 180, 7175–7183. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-Mcdermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Yang, X.Y.; Di Zheng, K.; Lin, K.; Zheng, G.; Zou, H.; Wang, J.M.; Lin, Y.Y.; Chuka, C.M.; Ge, R.S.; Zhai, W.; et al. Energy Metabolism Disorder as a Contributing Factor of Rheumatoid Arthritis: A Comparative Proteomic and Metabolomic Study. PLoS ONE 2015, 10, e0132695. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, J.; Xuan, J.; Jung, Y.H.; Cha, H.S.; Kim, K.H. Global Metabolite Profiling of Synovial Fluid for the Specific Diagnosis of Rheumatoid Arthritis from Other Inflammatory Arthritis. PLoS ONE 2014, 9, e97501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauridsen, M.B.; Bliddal, H.; Christensen, R.; Danneskiold-Samsøe, B.; Bennett, R.; Keun, H.; Lindon, J.C.; Nicholson, J.K.; Dorff, M.H.; Jaroszewski, J.W.; et al. 1H NMR Spectroscopy-Based Interventional Metabolic Phenotyping: A Cohort Study of Rheumatoid Arthritis Patients. J. Proteome Res. 2010, 9, 4545–4553. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Jiang, D.; Shao, J.; Sun, X. Magnetic molecularly imprinted polymer nanoparticles based electrochemical sensor for the measurement of Gram-negative bacterial quorum signaling molecules (N-acyl-homoserine-lactones). Biosens. Bioelectron. 2016, 75, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Jutley, G.S.; Sahota, K.; Sahbudin, I.; Filer, A.; Arayssi, T.; Young, S.P.; Raza, K. Relationship Between Inflammation and Metabolism in Patients With Newly Presenting Rheumatoid Arthritis. Front. Immunol. 2021, 12, 676105. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fang, K.; Wang, J.; Chang, X. Metabolomic analysis of synovial fluids from rheumatoid arthritis patients using quasi-targeted liquid chromatography-mass spectrometry/mass spectrometry. Clin. Exp. Rheumatol. 2021, 39, 1307–1315. [Google Scholar] [PubMed]
- Kim, J.; Kang, S.C.; Yoon, N.E.; Kim, Y.; Choi, J.; Park, N.; Jung, H.; Jung, B.H.; Ju, J.H. Metabolomic profiles of induced pluripotent stem cells derived from patients with rheumatoid arthritis and osteoarthritis. Stem Cell Res. Ther. 2019, 10, 319. [Google Scholar] [CrossRef]
- Moschen, A.R.; Gerner, R.R.; Tilg, H. Pre-B Cell Colony Enhancing Factor/NAMPT/Visfatin in Inflammation and Obesity-Related Disorders. Curr. Pharm. Des. 2010, 16, 1913–1920. [Google Scholar] [CrossRef]
- You, S.; Koh, J.H.; Leng, L.; Kim, W.U.; Bucala, R. The Tumor-Like Phenotype of Rheumatoid Synovium: Molecular Profiling and Prospects for Precision Medicine. Arthritis Rheumatol. 2018, 70, 637–652. [Google Scholar] [CrossRef]
- Bustamante, M.F.; Garcia-Carbonell, R.; Whisenant, K.D.; Guma, M. Fibroblast-like synoviocyte metabolism in the pathogenesis of rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 110. [Google Scholar] [CrossRef]
- Beckmann, J.; Schubert, J.; Morhenn, H.G.; Grau, V.; Schnettler, R.; Lips, K.S. Expression of choline and acetylcholine transporters in synovial tissue and cartilage of patients with rheumatoid arthritis and osteoarthritis. Cell Tissue Res. 2015, 359, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.M.; Yang, X.; Wang, H.; Saaoud, F.; Sun, Y.; Fong, D. The Microbial Metabolite Trimethylamine N-Oxide Links Vascular Dysfunctions and the Autoimmune Disease Rheumatoid Arthritis. Nutrients 2019, 11, 1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, R.; Smith, J.; Rocher-Ros, V.; Nadkarni, S.; Montero-Melendez, T.; D’Acquisto, F.; Bland, E.J.; Bombardieri, M.; Pitzalis, C.; Perretti, M.; et al. Lactate Regulates Metabolic and Pro-Inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol. 2015, 13, e1002202. [Google Scholar] [CrossRef] [PubMed]
- Michopoulos, F.; Karagianni, N.; Whalley, N.M.; Firth, M.A.; Nikolaou, C.; Wilson, I.D.; Critchlow, S.E.; Kollias, G.; Theodoridis, G.A. Targeted Metabolic Profiling of the Tg197 Mouse Model Reveals Itaconic Acid as a Marker of Rheumatoid Arthritis. J. Proteome Res. 2016, 15, 4579–4590. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, J.Y.; Liu, J.Q.; Yang, J.; Liu, Y.; Wang, C.; Ma, X.N.; Liu, B.L.; Xin, G.Z.; Liu, L.F. Succinate/NLRP3 Inflammasome Induces Synovial Fibroblast Activation: Therapeutical Effects of Clematichinenoside AR on Arthritis. Front. Immunol. 2016, 7, 532. [Google Scholar] [CrossRef] [Green Version]
- Surowiec, I.; Gjesdal, C.G.; Jonsson, G.; Norheim, K.B.; Lundstedt, T.; Trygg, J.; Omdal, R. Metabolomics study of fatigue in patients with rheumatoid arthritis naïve to biological treatment. Rheumatol. Int. 2016, 36, 703–711. [Google Scholar] [CrossRef]
- Madsen, R.K.; Lundstedt, T.; Gabrielsson, J.; Sennbro, C.J.; Alenius, G.M.; Moritz, T.; Rantapää-Dahlqvist, S.; Trygg, J. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res. Ther. 2011, 13, R19. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Carrio, J.; Alperi-López, M.; López, P.; Ballina-García, F.J.; Suárez, A. Non-Esterified Fatty Acids Profiling in Rheumatoid Arthritis: Associations with Clinical Features and Th1 Response. PLoS ONE 2016, 11, e0159573. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chen, J.; Hu, C.; Xie, Z.; Li, H.; Wei, S.; Wang, D.; Wen, C.; Xu, G. Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2016, 127, 60–67. [Google Scholar] [CrossRef]
- He, Z.; Liu, Z.; Gong, L. Biomarker identification and pathway analysis of rheumatoid arthritis based on metabolomics in combination with ingenuity pathway analysis. Proteomics 2021, 21, 2100037. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, T.; Feng, H.; Zhang, Y.; Li, L.; Zhao, A.; Niu, X.; Liang, F.; Wang, M.; Zhan, J.; et al. Serum Metabolic Signatures of Four Types of Human Arthritis. J. Proteome Res. 2013, 12, 3769–3779. [Google Scholar] [CrossRef]
- Krishna, V.; Yin, X.; Song, Q.; Walsh, A.; Pocalyko, D.; Bachman, K.; Anderson, I.; Madakamutil, L.; Nagpal, S. Integration of the Transcriptome and Genome-Wide Landscape of BRD2 and BRD4 Binding Motifs Identifies Key Superenhancer Genes and Reveals the Mechanism of Bet Inhibitor Action in Rheumatoid Arthritis Synovial Fibroblasts. J. Immunol. 2021, 206, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Che, N.; Xu, L.; Zhang, Q.; Wang, Q.; Tan, W.; Zhang, M. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin. Rheumatol. 2018, 37, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V. Urinary proteomics: Towards biomarker discovery, diagnostics and prognostics. Mol. Biosyst. 2008, 4, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Julià, A.; Vinaixa, M.; Domènech, E.; Fernández-Nebro, A.; Cañete, J.D.; Ferrándiz, C.; Tornero, J.; Gisbert, J.P.; Nos, P.; Casbas, A.G.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 133. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Rankin, K.N.; Wagner, J.; Fu, H.; Chang, D.; Weljie, A.M.; Saude, E.J.; Lix, B.; Adamko, D.J.; Shah, S.; et al. Investigations of the Effects of Gender, Diurnal Variation, and Age in Human Urinary Metabolomic Profiles. Anal. Chem. 2007, 79, 6995–7004. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The Apogee of the Omics Trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Teitsma, X.M.; Yang, W.; Jacobs, J.W.G.; Pethö-Schramm, A.; Borm, M.E.A.; Harms, A.C.; Hankemeier, T.; Van Laar, J.M.; Bijlsma, J.W.J.; Lafeber, F.P.J.G. Baseline Metabolic Profiles of Early Rheumatoid Arthritis Patients Achieving Sustained Drug-Free Remission after Initiating Treat-to-Target Tocilizumab, Methotrexate, or the Combination: Insights from Systems Biology 11 Medical and Health Sciences 1103 Clinical Sciences 06 Biological Sciences 0601 Biochemistry and Cell Biology. Arthritis Res. Ther. 2018, 20, 230. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, C.; Hiraishi, T.; Oku, T.; Okuma, K.; Suzumura, K.; Hashimoto, M.; Ito, H.; Aramori, I.; Hirayama, Y. Metabolomic approach to the exploration of biomarkers associated with disease activity in rheumatoid arthritis. PLoS ONE 2019, 14, e0219400. [Google Scholar] [CrossRef] [Green Version]
- Hur, B.; Gupta, V.K.; Huang, H.; Wright, K.A.; Warrington, K.J.; Taneja, V.; Davis, J.M.; Sung, J. Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity. Arthritis Res. Ther. 2021, 23, 164. [Google Scholar] [CrossRef]
- Ahn, J.K.; Kim, J.; Cheong, Y.E.; Kim, K.H.; Cha, H.-S. Variation in the synovial fluid metabolome according to disease activity of rheumatoid arthritis. Clin. Exp. Rheumatol. 2020, 38, 500–507. [Google Scholar]
- Figus, F.A.; Piga, M.; Azzolin, I.; McConnell, R.; Iagnocco, A. Rheumatoid arthritis: Extra-articular manifestations and comorbidities. Autoimmun. Rev. 2021, 20, 102776. [Google Scholar] [CrossRef] [PubMed]
- Young, S.P.; Nessim, M.; Falciani, F.; Trevino, V.; Banerjee, S.P.; Scott, R.A.; Murray, P.I.; Wallace, G.R. Metabolomic analysis of human vitreous humor differentiates ocular inflammatory disease. Mol. Vis. 2009, 15, 1210–1217. [Google Scholar] [PubMed]
- Shi, J.; Fan, J.; Su, Q.; Yang, Z. Cytokines and Abnormal Glucose and Lipid Metabolism. Front. Endocrinol. 2019, 10, 703. [Google Scholar] [CrossRef] [PubMed]
- Zabek, A.; Swierkot, J.; Malak, A.; Zawadzka, I.; Deja, S.; Bogunia-Kubik, K.; Mlynarz, P. Application of (1)H NMR-based serum metabolomic studies for monitoring female patients with rheumatoid arthritis. J. Pharm. Biomed. Anal. 2016, 117, 544–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priori, R.; Casadei, L.; Valerio, M.; Scrivo, R.; Valesini, G.; Manetti, C. 1H-NMR-Based Metabolomic Study for Identifying Serum Profiles Associated with the Response to Etanercept in Patients with Rheumatoid Arthritis. PLoS ONE 2015, 10, e0138537. [Google Scholar] [CrossRef]
- Kapoor, S.R.; Filer, A.; Fitzpatrick, M.A.; Fisher, B.A.; Taylor, P.C.; Buckley, C.D.; McInnes, I.B.; Raza, K.; Young, S.P. Metabolic Profiling Predicts Response to Anti-Tumor Necrosis Factor α Therapy in Patients with Rheumatoid Arthritis. Arthritis Rheum. 2013, 65, 1448–1456. [Google Scholar] [CrossRef]
- Tatar, Z.; Migne, C.; Petera, M.; Gaudin, P.; Lequerre, T.; Marotte, H.; Tebib, J.; Pujos Guillot, E.; Soubrier, M. Variations in the metabolome in response to disease activity of rheumatoid arthritis. BMC Musculoskelet. Disord. 2016, 17, 353. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Saegusa, J.; Onishi, A.; Morinobu, A. Biomarkers identified by serum metabolomic analysis to predict biologic treatment response in rheumatoid arthritis patients. Rheumatology 2019, 58, 2153–2161. [Google Scholar] [CrossRef]
- Urbaniak, B.; Plewa, S.; Klupczynska, A.; Sikorska, D.; Samborski, W.; Kokot, Z.J. Serum free amino acid levels in rheumatoid arthritis according to therapy and physical disability. Cytokine 2019, 113, 332–339. [Google Scholar] [CrossRef]
- Murillo-Saich, J.D.; Diaz-Torne, C.; Ortiz, M.A.; Coras, R.; Gil-Alabarse, P.; Pedersen, A.; Corominas, H.; Vidal, S.; Guma, M. Metabolomics profiling predicts outcome of tocilizumab in rheumatoid arthritis: An exploratory study. Metabolomics 2021, 17, 74. [Google Scholar] [CrossRef]
- Sweeney, S.R.; Kavanaugh, A.; Lodi, A.; Wang, B.; Boyle, D.; Tiziani, S.; Guma, M. Metabolomic profiling predicts outcome of rituximab therapy in rheumatoid arthritis. RMD Open 2016, 2, e000289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.K.; Chen, P.K.; Chen, C.C.; Chang, S.H.; Chen, C.H.; Chen, D.Y. Increased Levels of Omega-3 Fatty Acids and Dha Are Linked to Pain Reduction in Rheumatoid Arthritis Patients Treated with Janus Kinase Inhibitors. Nutrients 2021, 13, 3050. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Cuppen, B.V.J.; Welsing, P.M.J.; van Wietmarschen, H.; Harms, A.C.; Berger, R.; Koval, S.; Fritsch-Stork, R.D.E.; Bijlsma, J.W.J.; Hankemeier, T.; et al. Differences between serum polar lipid profiles of male and female rheumatoid arthritis patients in response to glucocorticoid treatment. Inflammopharmacology 2016, 24, 397–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artacho, A.; Isaac, S.; Nayak, R.; Flor-Duro, A.; Alexander, M.; Koo, I.; Manasson, J.; Smith, P.B.; Rosenthal, P.; Homsi, Y.; et al. The Pretreatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 931–942. [Google Scholar] [CrossRef]
- Wang, M.; Huang, J.; Fan, H.; He, D.; Zhao, S.; Shu, Y.; Li, H.; Liu, L.; Lu, S.; Xiao, C.; et al. Treatment of Rheumatoid Arthritis Using Combination of Methotrexate and Tripterygium Glycosides Tablets—A Quantitative Plasma Pharmacochemical and Pseudotargeted Metabolomic Approach. Front. Pharmacol. 2018, 9, 1051. [Google Scholar] [CrossRef] [Green Version]
- Radhakutty, A.; Mangelsdorf, B.L.; Drake, S.M.; Rowland, A.; Smith, M.D.; Mangoni, A.A.; Thompson, C.H.; Burt, M.G. Opposing effects of rheumatoid arthritis and low dose prednisolone on arginine metabolomics. Atherosclerosis 2017, 266, 190–195. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, T.; Wallman, J.K.; Jöud, A.; Schelin, M.E.C.; Ernestam, S.; van Vollenhoven, R.; Saevarsdottir, S.; Lampa, J. Pain Over Two Years After Start of Biologic Versus Conventional Combination Treatment in Early Rheumatoid Arthritis: Results From a Swedish Randomized Controlled Trial. Arthritis Care Res. 2021, 73, 1312–1321. [Google Scholar] [CrossRef]
- Chandrasekharan, U.M.; Wang, Z.; Wu, Y.; Wilson Tang, W.H.; Hazen, S.L.; Wang, S.; Elaine Husni, M. Elevated levels of plasma symmetric dimethylarginine and increased arginase activity as potential indicators of cardiovascular comorbidity in rheumatoid arthritis. Arthritis Res. Ther. 2018, 20, 123. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, H.; Oka, S.; Higuchi, T.; Shimada, K.; Hashimoto, A.; Matsui, T.; Tohma, S. Biomarkers for interstitial lung disease and acute-onset diffuse interstitial lung disease in rheumatoid arthritis. Ther. Adv. Musculoskelet. Dis. 2021, 13. [Google Scholar] [CrossRef]
- Alabarse, P.V.G.; Silva, J.M.S.; Santo, R.C.E.; Oliveira, M.S.; Almeida, A.S.; de Oliveira, M.S.; Immig, M.L.; Freitas, E.C.; Teixeira, V.O.N.; Bathurst, C.L.; et al. Metabolomic Biomarker Candidates for Skeletal Muscle Loss in the Collagen-Induced Arthritis (Cia) Model. J. Pers. Med. 2021, 11, 837. [Google Scholar] [CrossRef]




| Metabolite | Source | Mechanism of Action | Identified Condition | References |
|---|---|---|---|---|
| TMAO | Gut microbial metabolism from dietary choline and phosphatidylcholine (lecithin) | Increase glucose tolerance, inhibits hepatic insulin signaling and promotes adipose tissue inflammation | Increase in chronic kidney disease, type-2 diabetes mellitus, atherosclerosis | [32,33,34,35,36] |
| BCAA’s | Diet consumption of meat, dairy and vegetables | Induced NADPH inflammation and Akt/mTOR signaling, as well as promoting pro-inflammatory cytokines (IL-6, TNF) and blood of peripheral mononuclear cells by diet | Increase in: maple syrup urine disease, heart, kidney and spleen hypertrophy type 2 diabetes | [41,42,43] |
| Glutamine | Mainly synthesized by the GS and hydrolyzed by GLS. | Promotes enterocyte proliferation, regulates tight junction proteins, suppresses pro-inflammatory signaling pathways and protects cells against apoptosis and cellular stresses during normal and pathologic conditions | Trauma, sepsis, inflammatory bowel diseases and cardiovascular diseases | [44,45,46,47] |
| Succinate | TCA | Stabilizes transcription factor HIF-1a in tumors and in activated macrophages. Stimulates dendritic cells via its receptor succinate receptor1. | Peritonitis, cancer, diabetic and metabolic disease rodent models | [24,25,30,31] |
| Itaconate | TCA | Targets on ATF3-IκBζ pathway in a Nrf2-independent manner to mediate the inflammatory response. | Reperfusion injury, inflammatory disease and bacterial infections | [26,27,28,29] |
| Oxylipins | Oxygenation PUFAs: AA and LA | Activate PPARs or through GPCRs | Hyperlipidemia, hypertension, thrombosis, hemostasis and diabetes | [48,49,50] |
| SCFA | Products of dietary fiber metabolism by the gut microbiome | Activate FFA2 and FFA3 receptors and GPR109A through the inhibition of HDACs. | Salmonella infection, Eczema and Alzheimer’s disease | [51,52,53] |
| BA | BAs are synthesized in the liver and released into the gastrointestinal tract to aid in lipid digestion | Suppressed the production of LPS-induced inflammatory cytokines in macrophages | Insulin resistance | [54] |
| IDO | Tryptophan products | Toxic to T cells and induce cell death by apoptosis | Alzheimer’s disease, multiple sclerosis, Huntington’s disease and Human Lymphocyte Antigen-G | [40,55,56,57] |
| FFA | Derived from alpha-linolenic acid-omega-3-and linoleic acid-omega-6 or synthesized in the body. | Binding to cell-surface receptors of the GPCR family and regulated energy homeostasis indirectly via hormonal signaling | Type-2 Diabetes Colorectal cancer Systemic Lupus Erythematosus | [58,59] |
| Source | Treatment Use | Method Applied | Metabolites | Reference |
|---|---|---|---|---|
| Urine | TNFi | GC/TOF MS | Histamine, glutamine, phenylacetic acid, xanthine, xanthurenic acid and creatinine were upregulated in urine samples from patients who had a good response to TNF therapy, while ethanolamine, hydroxyphenylpyruvic acid and phosphocreatine were downregulated. | [136] |
| Serum | DMARDS: MTX or leflunomide; bDMARDS: TNFi | HPLC-MS/MS | Threonine: Distinction of RA patients treated with MTX/leflunomide vs. infliximab/adalimumab/etanercept/tocilizumab and infliximab/adalimumab/etanercept/tocilizumab-prednisolone/NSAID Tryptophan: differentiated RA patients treated with methotrexate/leflunomide- vs. infliximab/adalimumab/etanercept/tocilizumab. | [139] |
| Serum | Etanercept | 1H NMR | Increase in isoleucine, leucine, valine, alanine, glutamine, tyrosine and glucose levels and a decrease in 3-hydroxybutyrate levels N Etanercept good responders | [135] |
| Blood | Infliximab, abatacept or etanercept. | RP-UHPLC ESI-QTOF-MS | Two different metabolic profiles splitting good responders from non-responders: Carbohydrate derivatives (D-glucose, D-fructose, sucrose and maltose) | [137] |
| Plasma | Tocilizumab | H-NMR | Concentrations of 3-hydroxybutyrate and phenylalanine improved the ability to specifically predict TCZ responders | [140] |
| Serum | Rituximab | NMR-MS | Phosphatidylethanolamines, phosphatidyserines and phosphatidylglycerols were downregulated in responders; 37 lipids were different between responder and non-responders. | [141] |
| Serum | TNFi | CE-TOFMS | Association with TNFi: Betonicine, glycerol 3-phosphate, N-acetylalanine, hexanoic acid and taurine are associated with the response to TNFi in RA. Associated with Abatacept: Citric acid, quinic acid and 3-aminobutyric acid. | [138] |
| Serum | Tocilizumab | MS | Changes in arachidonic acid metabolism | [127] |
| Serum | Etanercept/adalimumab | 1H NMR | 3-hydroxyisobutyrate, lysine, L5, acetoacetate, creatine, GPC+APC, histidine and phenylalanine were elevated in RA, whereas leucine, acetate, betaine and formate were lower. | [134] |
| Serum | Tofacitinib/baricitinib | 1H-NMR | Levels of omega-3 fatty acids DHA were increased in JAKi-treated patients. DHA was associated with decreases in pain. | [142] |
| Serum | GC | LC-MS/MS | Elevated lysophosphatidylcholines and lysophosphatidylethanolamines in women. | [143] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartikoski, B.J.; De Oliveira, M.S.; Do Espírito Santo, R.C.; Dos Santos, L.P.; Dos Santos, N.G.; Xavier, R.M. A Review of Metabolomic Profiling in Rheumatoid Arthritis: Bringing New Insights in Disease Pathogenesis, Treatment and Comorbidities. Metabolites 2022, 12, 394. https://doi.org/10.3390/metabo12050394
Bartikoski BJ, De Oliveira MS, Do Espírito Santo RC, Dos Santos LP, Dos Santos NG, Xavier RM. A Review of Metabolomic Profiling in Rheumatoid Arthritis: Bringing New Insights in Disease Pathogenesis, Treatment and Comorbidities. Metabolites. 2022; 12(5):394. https://doi.org/10.3390/metabo12050394
Chicago/Turabian StyleBartikoski, Bárbara Jonson, Marianne Schrader De Oliveira, Rafaela Cavalheiro Do Espírito Santo, Leonardo Peterson Dos Santos, Natália Garcia Dos Santos, and Ricardo Machado Xavier. 2022. "A Review of Metabolomic Profiling in Rheumatoid Arthritis: Bringing New Insights in Disease Pathogenesis, Treatment and Comorbidities" Metabolites 12, no. 5: 394. https://doi.org/10.3390/metabo12050394