Effect of Denoising and Deblurring 18F-Fluorodeoxyglucose Positron Emission Tomography Images on a Deep Learning Model’s Classification Performance for Alzheimer’s Disease
Abstract
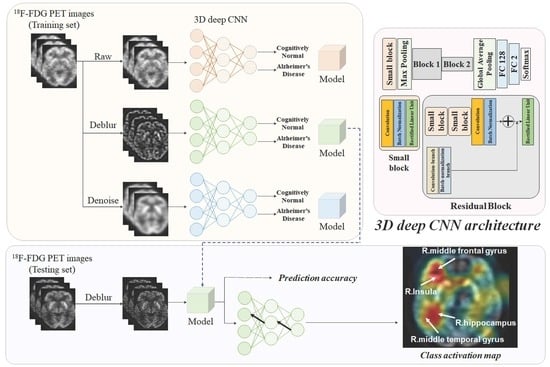
:1. Introduction
2. Results
2.1. Demographic Characteristics and Clinical Assessments
2.2. Classification Performance
2.3. Activation Maps Associated with the Classification of Alzheimer’s Disease and Cognitively Normal Controls
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Preprocessing
4.2. Cognitive Assessment
4.3. Image Restoration
4.4. Proposed Framework
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngamsombat, C.; Dechasasawat, T.; Wongsripuemtet, J.; Charnchaowanish, P.; Muangpaisan, W.; Chawalparit, O. The evaluation of posterior cingulate gyrus by diffusion tensor imaging in Alzheimer’s disease patients compared with normal control subjects. Siriraj Med. J. 2019, 71, 117–122. [Google Scholar]
- Nordberg, A.; Rinne, J.O.; Kadir, A.; Långström, B. The use of pet in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R.; Knopman, D.S.; Jagust, W.J.; Shaw, L.M.; Aisen, P.S.; Weiner, M.W.; Petersen, R.C.; Trojanowski, J.Q. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010, 9, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Forlenza, O.V.; Radanovic, M.; Talib, L.L.; Aprahamian, I.; Biniz, B.S.; Zetterberg, H.; Gattaz, W.F. Cerebrospinal fluid biomarkers in Alzheimer’s disease: Diagnostic accuracy and prediction of dementia. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2015, 1, 455–463. [Google Scholar] [CrossRef]
- Ding, Y.; Sohn, J.H.; Kawczynski, M.G.; Trivedi, H.; Harnish, R.; Jenkins, N.W.; Lituiev, D.; Copeland, T.P.; Aboian, M.S.; Aparici, C.M.; et al. A deep learning model to predict a diagnosis of alzheimer disease by using 18F-FDG PET of the brain. Radiology 2019, 290, 456–464. [Google Scholar] [CrossRef]
- Long, X.; Chen, L.; Jiang, C.; Zhang, L.; Alzheimer’s Disease Neuroimaging Initiative. Prediction and classification of alzheimer disease based on quantification of MRI deformation. PLoS ONE 2017, 12, e0173372. [Google Scholar] [CrossRef]
- Bohen, N.I.; Djang, D.S.W.; Herholz, K.; Anzai, Y.; Minoshima, S. Effectiveness and safety of 18F-FDG PET in the evaluation of dementia: A review of the recent literature. J. Nucl. Med. 2012, 53, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Zeng, R.; Yu, L.; Feng, Y.; Chen, C.; Li, F.; Liu, Y.; Huang, Y.; Huang, Z.; Alzheimer’s Disease Neuroimaging Initiative. Deep-learning radiomics for discrimination conversion of Alzheimer’s disease in patients with mild cognitive impairment: A study based on 18F-FDG PET imaging. Front. Aging Neurosci. 2021, 13, 764872. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, Z. The risk prediction of Alzheimer’s disease based on the deep learning model of 18F-FDG positron emission tomography. Saudi J. Biol. Sci. 2020, 27, 659–665. [Google Scholar] [CrossRef]
- Röhrbein, F.; Goddard, P.; Schneider, M.; James, G.; Guo, K. How does image noise affect actual and predicted human gaze allocation in assessing image quality? Vis. Res. 2015, 112, 11–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodge, S.; Karam, L. Understanding how image quality affects deep neural networks. In Proceedings of the 2016 Eighth International Conference on Quality of Multimedia Experience (QoMEX), Lisbon, Portugal, 6–8 June 2016; pp. 1–6. [Google Scholar]
- Christensen, A.N. PET and PET/CT physics. In PET/CT Atlas on Quality Control and Image Artefacts; IAEA Human Health Series; International Atomic Energy Agency: Vienna, Austria, 2014; pp. 10–27. [Google Scholar]
- Shi, H.; Li, H.; Zhang, D.; Cheng, C.; Cao, X. An efficient feature generation approach based on deep learning and feature selection techniques for traffic classification. Comput. Netw. 2018, 132, 81–98. [Google Scholar] [CrossRef]
- Kwak, N.; Choi, C.-H. Input feature selection for classification problems. IEEE Trans. Neural Netw. 2002, 13, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Luo, S.; Chiong, R. Introducing transfer learning to 3D ResNet-18 for Alzheimer’s disease detection on MRI images. In Proceedings of the 2020 35th International Conference on Image and Vision Computing New Zealand (IVCNZ), Wellington, New Zealand, 25–27 November 2020. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Nazaré, T.S.; da Costa, G.B.P.; Contato, W.A.; Ponti, M. Deep convolutional neural networks and noisy images. In Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications; Springer: Cham, Switzerland, 2018; pp. 416–424. [Google Scholar]
- da Costa, G.B.P.; Contato, W.A.; Nazaré, T.S.; Neto, J.E.S.B.; Ponti, M. An empirical study on the effects of different types of noise in image classification tasks. arXiv 2016, arXiv:1609.02781. [Google Scholar]
- Song, T.A.; Yang, F.; Chowdhury, S.R.; Kim, K.; Johnson, K.A.; Fakhri, G.E.; Li, Q.; Dutta, J. Pet image deblurring and super-resolution with and MR-based joint entropy prior. IEEE Trans. Comput. Imaging 2019, 5, 530–539. [Google Scholar] [CrossRef]
- Cui, J.; Gong, K.; Guo, N.; Wu, C.; Meng, X.; Kim, K.; Zheng, K.; Wu, Z.; Fu, L.; Xu, B.; et al. PET image denoising using unsupervised deep learning. Eur. J. Nucl. Med. Mol. Imaging 2021, 46, 2780–2789. [Google Scholar] [CrossRef]
- Song, T.-A.; Yang, F.; Dutta, J. Noise2void: Unsupervised denoising of PET images. Phys. Med. Biol. 2021, 66, 214002. [Google Scholar] [CrossRef]
- Liu, J.; Malekzadeh, M.; Mirian, N.; Song, T.-A.; Liu, C.; Dutta, J. Artificial intelligence-based image enhancement in PET imaging. PET Clin. 2021, 16, 553–576. [Google Scholar] [CrossRef]
- Joshi, A.; Koeppe, R.A.; Fessler, J.A. Reducing between scanner differences in multi-center PET studies. Neuroimage 2009, 46, 154–159. [Google Scholar] [CrossRef] [Green Version]
- Mishra, B.K.; Thakker, D.; Mazumdar, S.; Neagu, D.; Gheorghe, M.; Simpson, S. A novel application of deep learning with image cropping: A smart city use case for flood monitoring. J. Reliab. Intell. Environ. 2020, 6, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chen, X.; Zheng, W.; Xia, M.; Han, Y.; Song, H.; Li, K.; He, Y.; Wang, Z. Altered functional connectivity of insular subregions in Alzheimer’s disease. Front. Aging Neurosci. 2018, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Guo, H.; Gao, Y.; Wang, X.; Cui, H.; Chen, Z.; Wang, B.; Xiang, J. Altered directed functional connectivity of the hippocampus in mild cognitive impairment and Alzheimer’s disease: A resting-state fMRI study. Front. Aging Neurosci. 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trzepacz, P.T.; Yu, P.; Bhamidipati, P.K.; Willis, B.; Forrester, T.; Tabas, L.; Schwarz, A.J.; Saykin, A.J.; Alzheimer’s Disease Neuroimaging Initiative. Frontolimbic atrophy is associated with agitation and aggression in mild cognitive impairment and Alzheimer’s disease. Alzheimer’s Dement. 2013, 9, S95–S104. [Google Scholar] [CrossRef] [Green Version]
- Marcus, C.; Mena, E.; Subramaniam, R.M. Brain PET in the diagnosis of Alzheimer’s disease. Clin. Nucl. Med. 2014, 39, e413–e426. [Google Scholar] [CrossRef] [Green Version]
- Mosconi, L.; De Santi, S.; Li, J.; Tsui, W.H.; Li, Y.; Boppana, M.; Laska, E.; Rusinek, H.; de Leon, M.J. Hippocampal hypometabolism predicts cognitive decline from normal aging. Neurobiol. Aging 2008, 29, 676–692. [Google Scholar] [CrossRef] [Green Version]
- Peters, F.; Collette, F.; Degueldre, C.; Sterpenich, V.; Majerus, S.; Salmon, E. The neural correlates of verbal short-term memory in Alzheimer’s disease: An fMRI study. Brain 2009, 132, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Silverman, D.H.; Small, G.W.; Chang, C.Y.; Lu, C.S.; Kung De Aburto, M.A.; Chen, W.; Czernin, J.; Rapoport, S.I.; Pietrini, P.; Alexander, G.E.; et al. Positron emission tomography in evaluation of dementia: Regional brain metabolism and long-term outcome. JAMA 2001, 286, 2120–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, Y.-N.; Xu, W.; Li, J.-Q.; Guo, Y.; Cui, M.; Chen, K.-L.; Huang, Y.-Y.; Dong, Q.; Tan, L.; Yu, J.-T.; et al. FDG-PET as an independent biomarker for Alzheimer’s biological diagnosis: A longitudinal study. Alzheimer’s Res. Ther. 2019, 11, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.-P.; Sweeney, E.M.; Muschelli, J.; Crainiceanu, C.P.; Shinohara, R.T.; Alzheimer’s Disease Neuroimaging Initiative. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage 2016, 132, 198–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torbati, M.E.; Minhas, D.S.; Ahmad, G.; O’Connor, E.E.; Muschelli, J.; Laymon, C.M.; Yang, Z.; Cohen, A.D.; Aizenstein, H.J.; Klunk, W.E.; et al. A multi-scanner neuroimaging data harmonization using RAVEL and ComBat. Neuroimage 2021, 245, 118703. [Google Scholar] [CrossRef]
- Mosteller, F.; Turkey, J. Data analysis, including statistics. In The Handbook of Social Psychology; Gardner, L., Eliot, A., Eds.; Springer & Addison-Wesley: Reading, MA, USA, 1968; pp. 109–112. [Google Scholar]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer’s disease. Neurology 1984, 37, 939–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouter, C.; Henniges, P.; Franke, T.N.; Irwin, C.; Sahlmann, C.O.; Sichler, M.E.; Beindorff, N.; Bayer, T.A.; Bouter, Y. 18F-FDG-PET detects drastic changes in brain metabolism in the Tg4–42 model of Alzheimer’s disease. Front. Aging Neurosci. 2019, 10, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolls, E.T.; Joliot, M.; Tzourio-Mazoyer, N. Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage 2015, 122, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Hughes, C.P.; Berg, L.; Danziger, W.L.; Coben, L.A.; Martin, R.L. A new clinical scale for the staging of dementia. Br. J. Psychiatry 1982, 140, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.C. The clinical dementia rating (CDR): Current vision and scoring rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Waring, S.C.; Cullum, C.M.; Hall, J.; Lacritz, L.; Massman, P.J.; Lupo, P.J.; Reisch, J.S.; Doody, R.; Texas Alzheimer’s Research Consortium. Staging dementia using clinical dementia rating scale sum of boxes scores. Arch. Neurol. 2008, 65, 1091–1095. [Google Scholar] [CrossRef] [Green Version]
- da Silva, E.A.B.; Mendonça, G.V. The Electrical Engineering Handbook, 4-Digital Image Processing; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Wang, Y.; Yang, J.; Yin, W.; Zhang, Y. A new alternating minimization algorithm for total variation image reconstruction. SIAM J. Imaging Sci. 2008, 1, 248–272. [Google Scholar] [CrossRef]
- Yang, J.; Yin, W.; Zhang, Y.; Wang, Y. A fast algorithm for edge-preserving variational multichannel image restoration. SIAM J. Imaging Sci. 2009, 2, 569–592. [Google Scholar] [CrossRef]
- Ren, D.; Zuo, W.; Zhang, D.; Xu, J.; Zhang, L. Partial deconvolution with inaccurate blur kernel. IEEE Trans. Image Process. 2018, 27, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Fessler, J.A. A splitting-based iterative algorithm for accelerated statistical X-ray CT reconstruction. IEEE Trans. Med. Imaging 2012, 31, 677–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannistraci, C.V.; Abbas, A.; Gao, X. Median modified wiener filter for nonlinear adaptive spatial denoising of protein NMR multidimensional spectra. Sci. Rep. 2015, 5, 8017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.R.; Kang, S.H.; Lee, Y. Median modified wiener filter for improving the image quality of gamma camera images. Nucl. Eng. Technol. 2020, 52, 2328–2333. [Google Scholar] [CrossRef]
- Xu, G.; Wu, H.Z.; Shi, Y.Q. Ensemble of CNNs for steganalysis: An empirical study. In Proceedings of the 4th ACM Workshop on Information Hiding and Multimedia Security, Galicia, Spain, 20–22 June 2016; pp. 103–107. [Google Scholar]
- Ting, M.K. Confusion matrix. In Encyclopedia of Machine Learning and Data Mining; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2017. [Google Scholar]
- Chicco, D.; Tötsch, N.; Jurman, G. The Matthews Correlation Coefficient (MCC) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. Methodology 2021, 14, 13. [Google Scholar] [CrossRef]





| Cognitively Normal Controls (n = 155) | Patients with Alzheimer’s Disease (n = 66) | p | |
|---|---|---|---|
| Age | 75.31 ± 6.57 | 74.44 ± 8.39 | 0.412 |
| Male sex | 81 (52.26) | 33 (50.00) | 0.759 |
| Education | 16.17 ± 2.89 | 15.35 ± 2.90 | 0.054 |
| APOE ε4, carriers | 43 (27.74) | 45 (68.18) | <0.001 |
| MMSE score | 29.03 ± 1.21 | 23.26 ± 2.15 | <0.001 |
| Global CDR score | 0.00 ± 0.00 | 0.80 ± 0.29 | <0.001 |
| CDR Sum of Boxes | 0.04 ± 0.15 | 4.53 ± 1.67 | <0.001 |
| Memory | 0.00 ± 0.00 | 1.05 ± 0.40 | <0.001 |
| Orientation | 0.00 ± 0.00 | 0.89 ± 0.40 | <0.001 |
| Judgment | 0.04 ± 0.13 | 0.87 ± 0.34 | <0.001 |
| Community affairs | 0.01 ± 0.08 | 0.76 ± 0.49 | <0.001 |
| Hobbies | 0.00 ± 0.00 | 0.73 ± 0.51 | <0.001 |
| Personal care | 0.00 ± 0.00 | 0.23 ± 0.42 | <0.001 |
| Sensitivity | Specificity | Accuracy | F1-Score | MCC | |
|---|---|---|---|---|---|
| Raw 18F-FDG PET | 0.72 ± 0.07 | 0.32 ± 0.08 | 0.60 ± 0.07 | 0.71 ± 0.05 | 0.10 ± 0.09 |
| Deblurred 18F-FDG PET (σb = 1) | 0.83 ± 0.07 | 0.30 ± 0.09 | 0.67 ± 0.03 | 0.78 ± 0.03 | 0.14 ± 0.04 |
| Deblurred 18F-FDG PET (σb = 2) | 0.91 ± 0.08 | 0.25 ± 0.15 | 0.71 ± 0.05 | 0.81 ± 0.04 | 0.21 ± 0.15 |
| Denoised 18F-FDG PET (σn = 3) | 0.75 ± 0.08 | 0.65 ± 0.05 | 0.72 ± 0.05 | 0.79 ± 0.05 | 0.39 ± 0.08 |
| Denoised 18F-FDG PET (σn = 5) | 0.85 ± 0.06 | 0.48 ± 0.13 | 0.74 ± 0.05 | 0.82 ± 0.04 | 0.35 ± 0.14 |
| Sensitivity | Specificity | Accuracy | F1-Score | MCC | |
|---|---|---|---|---|---|
| Raw 18F-FDG PET | 0.78 ± 0.06 | 0.63 ± 0.13 | 0.74 ± 0.03 | 0.81 ± 0.03 | 0.40 ± 0.09 |
| Deblurred 18F-FDG PET (σb = 1) | 0.85 ± 0.06 | 0.68 ± 0.06 | 0.80 ± 0.05 | 0.85 ± 0.04 | 0.53 ± 0.10 |
| Deblurred 18F-FDG PET (σb = 2) | 0.89 ± 0.06 | 0.67 ± 0.10 | 0.82 ± 0.07 | 0.88 ± 0.05 | 0.57 ± 0.17 |
| Denoised 18F-FDG PET (σn = 3) | 0.83 ± 0.08 | 0.50 ± 0.13 | 0.73 ± 0.08 | 0.81 ± 0.05 | 0.34 ± 0.19 |
| Denoised 18F-FDG PET (σn = 5) | 0.88 ± 0.06 | 0.63 ± 0.08 | 0.80 ± 0.03 | 0.86 ± 0.02 | 0.53 ± 0.05 |
| Scanner Model | Scintillator Materials | Scintillator Size (mm3) | Reconstruction Algorithm | Image Size | Slice Thickness (mm) |
|---|---|---|---|---|---|
| Siemens HRRT | LSO | 2.1 × 2.1 × 20 | OSEM-3D | 256 × 256 × 207 | 1.2 |
| Siemens HR+ | BGO | 4.05 × 4.39 × 30 | FORE/OSEM-2D | 128 × 128 × 63 | 2.4 |
| Siemens Accel | LSO | 6.45 × 6.45 × 25 | FORE/OSEM-2D | 128 × 128 × 47 | 3.4 |
| Siemens Exact | BGO | 6.75 × 6.75 × 20 | FORE/OSEM-2D | 128 × 128 × 47 | 3.4 |
| Siemens SOMATOM Definition AS mCT | LSO | 4.0 × 4.0 × 20 | OSEM-3D | 400 × 400 × 109 | 2.0 |
| Siemens SOMATOM Definition AS mCT | LSO | 4.0 × 4.0 × 20 | OSEM-3D | 400 × 400 × 81 | 2.0 |
| Siemens Biograph 64 | LSO | 4.0 × 4.0 × 20 | OSEM-3D | 400 × 400 × 109 | 2.0 |
| Siemens Biograph mCT 20 | LSO | 4.0 × 4.0 × 20 | OSEM-3D | 256 × 256 × 81 | 2.0 |
| Siemens 1094 | LSO | 4.0 × 4.0 × 20 | OSEM-3D | 336 × 336 × 109 | 2.0 |
| Layer | Shape | Filter of Pooling | Stride/Padding |
|---|---|---|---|
| Input | 128 × 128 × 79 × 1 or 128 × 12 × 810 × 1 | - | - |
| Conv | 64 × 64 × 40 × 64 or 64 × 64 × 5 × 64 | 7 × 7 × 7 | 2/3 |
| BN | - | - | |
| ReLU | - | - | |
| Max pooling | 32 × 32 × 20 × 64 or 32 × 32 × 3 × 64 | 3 × 3 × 3 | 2/1 |
| Conv and Conv-branch | 32 × 32 × 20 × 64 or 32 × 32 × 3 × 64 | 3 × 3 × 3 | 1/1 |
| BN and BN-branch | - | - | |
| ReLU | - | - | |
| Conv and Conv-branch | 16 × 16 × 10 × 128 or 16 × 16 × 2 × 128 | 3 × 3 × 3 | 1/1 |
| BN and BN-branch | - | - | |
| ReLU | - | - | |
| Global average pooling | 1 × 1 × 1 × 128 | - | - |
| Fully connected-128 | 1 × 1 × 1 × 128 | - | - |
| Fully connected-2 | 1 × 1 × 1 × 2 | - | - |
| Softmax | 1 × 1 × 1 × 2 | - | - |
| Classification output | 1 × 1 × 1 × 2 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-H.; Yun, C.-S.; Kim, K.; Lee, Y. Effect of Denoising and Deblurring 18F-Fluorodeoxyglucose Positron Emission Tomography Images on a Deep Learning Model’s Classification Performance for Alzheimer’s Disease. Metabolites 2022, 12, 231. https://doi.org/10.3390/metabo12030231
Lee M-H, Yun C-S, Kim K, Lee Y. Effect of Denoising and Deblurring 18F-Fluorodeoxyglucose Positron Emission Tomography Images on a Deep Learning Model’s Classification Performance for Alzheimer’s Disease. Metabolites. 2022; 12(3):231. https://doi.org/10.3390/metabo12030231
Chicago/Turabian StyleLee, Min-Hee, Chang-Soo Yun, Kyuseok Kim, and Youngjin Lee. 2022. "Effect of Denoising and Deblurring 18F-Fluorodeoxyglucose Positron Emission Tomography Images on a Deep Learning Model’s Classification Performance for Alzheimer’s Disease" Metabolites 12, no. 3: 231. https://doi.org/10.3390/metabo12030231