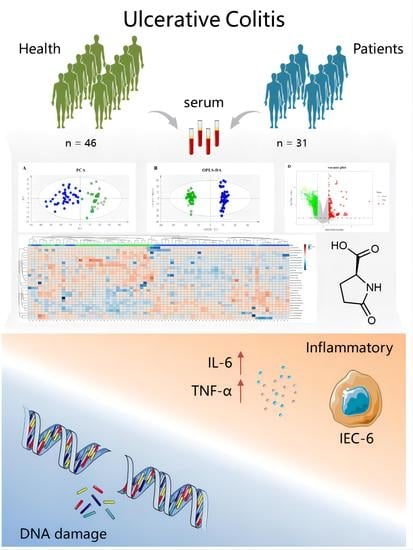
Discovery and Validation of Potential Serum Biomarkers with Pro-Inflammatory and DNA Damage Activities in Ulcerative Colitis: A Comprehensive Untargeted Metabolomic Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population and Study Design
2.2. Preparation of the Samples for Metabolomics Extraction
2.3. HPLC-Q-TOF MS Analysis
2.4. Method Assessment
2.5. Data Processing and Statistical Analysis
2.6. Cell Culture
2.7. Analysis of mRNA Levels by Quantitative Real-Time PCR (qPCR)
2.8. Western Blot Analysis
2.9. Immunofluorescence Detection of γH2AX
2.10. Comet Assay
2.11. Statistical Analysis
3. Results
3.1. Basic Characteristics of the Participants
3.2. Multivariate Statistical Analysis of Potential Biomarkers for UC
3.3. Enrichment Analysis of Metabolic Pathway and Regulatory Enzymes
3.4. Validation of Potential Pro-Inflammatory and DNA Damage Activity of Pyroglutamic Acid
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piovani, D.; Danese, S.; Peyrin-Biroulet, L.; Nikolopoulos, G.K.; Lytras, T.; Bonovas, S. Environmental risk factors for inflammatory bowel diseases: An umbrella review of meta-analyses. Gastroenterology 2019, 157, 647–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Prim. 2020, 6, 74–94. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.; Mehandru, S.; Patrick, B.; Peyrin-Biroulet, L.; Colombel, J.F. Ulcerative colitis. Lancet 2016, 389, 1756–1770. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Feagan, B.G.; Wolf, D.C.; D’Haens, G.; Vermeire, S.; Hanauer, S.B.; Ghosh, S.; Smith, H.; Cravets, M.; Frohna, P.A.; et al. Ozanimod induction and maintenance treatment for ulcerative colitis. New Engl. J. Med. 2016, 374, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Chang, T.A.; Chao, T.H.; Chen, J.S.; Chou, J.W.; Chou, Y.H.; Chuang, C.H.; Hsu, W.H.; Huang, T.Y.; Hsu, T.C.; et al. Management of ulcerative colitis in Taiwan: Consensus guideline of the Taiwan Society of Inflammatory Bowel Disease. Intest. Res. 2017, 15, 266–284. [Google Scholar] [CrossRef] [PubMed]
- Nakase, H.; Uchino, M.; Shinzaki, S.; Matsuura, M.; Matsuoka, K.; Kobayashi, T.; Saruta, M.; Hirai, F.; Hata, K.; Hiraoka, S.; et al. Evidence-based clinical practice guidelines for inflammatory bowel disease 2020. J. Gastroenterol. 2021, 56, 489–526. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Itzkowitz, S.H. Colorectal cancer in inflammatory bowel disease: Mechanisms and management. Gastroenterology 2022, 162, 715–730. [Google Scholar] [CrossRef]
- Krugliak, C.N.; Torres, J.; Rubin, D.T. What does disease progression look like in ulcerative colitis, and how might it be prevented? Gastroenterology 2022, 162, 1396–1408. [Google Scholar] [CrossRef]
- Ge, C.; Lu, Y.; Shen, H.; Zhu, L. Monitoring of intestinal inflammation and prediction of recurrence in ulcerative colitis. Scand. J. Gastroenterol. 2022, 57, 513–524. [Google Scholar] [CrossRef]
- Krzystek-Korpacka, M.; Kempiński, R.; Bromke, M.; Neubauer, K. Biochemical biomarkers of mucosal healing for inflammatory bowel disease in adults. Diagnostics 2020, 10, 367. [Google Scholar] [CrossRef]
- Van, R.; Van, V.; Fidler, V. Faecal calprotectin for screening of patients with suspected inflammatory bowel disease: Diagnostic meta-analysis. BMJ 2010, 341, 3369. [Google Scholar]
- Tibble, J.A.; Sigthorsson, G.; Foster, R.; Forgacs, I.; Bjarnason, I. Use of surrogate markers of inflammation and Rome criteria to distinguish organic from nonorganic intestinal disease. Gastroenterology 2002, 123, 450–460. [Google Scholar] [CrossRef] [Green Version]
- Alarcon-Barrera, J.C.; Kostidis, S.; Ondo-Mendez, A.; Giera, M. Recent advances in metabolomics analysis for early drug development. Drug Discov. Today 2022, 27, 1763–1773. [Google Scholar] [CrossRef]
- Sandlers, Y. The future perspective: Metabolomics in laboratory medicine for inborn errors of metabolism. Transl. Res. 2017, 189, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Del Real, A.; Ciordia, S.; Sañudo, C.; Garcia-Ibarbia, C.; Roa-Bautista, A.; Ocejo-Viñals, J.G.; Corrales, F.; Riancho, J.A. Analysis of serum proteome after treatment of osteoporosis with anabolic or antiresorptive drugs. Metabolites 2022, 12, 399–410. [Google Scholar] [CrossRef]
- Tsamouri, M.M.; Durbin-Johnson, B.P.; Culp, W.T.N.; Palm, C.A.; Parikh, M.; Kent, M.S.; Ghosh, P.M. Untargeted metabolomics identify a panel of urinary biomarkers for the diagnosis of urothelial carcinoma of the bladder, as compared to urolithiasis with or without urinary tract infection in dogs. Metabolites 2022, 12, 200–221. [Google Scholar] [CrossRef]
- Homorogan, C.; Nitusca, D.; Enatescu, V.; Schubart, P.; Moraru, C.; Socaciu, C.; Marian, C. Untargeted plasma metabolomic profiling in patients with major depressive disorder using ultra-high performance liquid chromatography coupled with mass spectrometry. Metabolites 2021, 11, 466–480. [Google Scholar] [CrossRef]
- Alsoud, L.O.; Soares, N.C.; Al-Hroub, H.M.; Mousa, M.; Kasabri, V.; Bulatova, N.; Suyagh, M.; Alzoubi, K.H.; El-Huneidi, W.; Abu-Irmaileh, B.; et al. Identification of Insulin Resistance Biomarkers in Metabolic Syndrome Detected by UHPLC-ESI-QTOF-MS. Metabolites 2022, 12, 508–527. [Google Scholar] [CrossRef]
- Le Cosquer, G.; Buscail, E.; Gilletta, C.; Deraison, C.; Duffas, J.P.; Bournet, B.; Tuyeras, G.; Vergnolle, N.; Buscail, L. Incidence and risk factors of cancer in the anal transitional zone and ileal pouch following surgery for ulcerative colitis and familial adenomatous polyposis. Cancers 2022, 14, 530. [Google Scholar] [CrossRef]
- Alexeev, E. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [Green Version]
- Lamas, B.; Natividad, J.; Sokol, H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal. Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef]
- Michael, P.; Ellen, A.; Ute, F. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar]
- Teresa, Z.; Rossana, G.; Cristina, C. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar]
- Jahidul, I.; Shoko, S.; Kouichi, W. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017, 42, 43–50. [Google Scholar]
- Kim, C.J.; Kovacs-Nolan, J.A.; Yang, C.; Archbold, T.; Fan, M.Z.; Mine, Y. L-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J. Nutr. Biochem. 2010, 21, 468–475. [Google Scholar] [CrossRef]
- Qi, Y.; Jiang, C.; Tanaka, N.; Krausz, K.W.; Brocker, C.N.; Fang, Z.Z.; Bredell, B.X.; Shah, Y.M.; Gonzalez, F.J. PPARα-dependent exacerbation of experimental colitis by the hypolipidemic drug fenofibrate. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, 564–573. [Google Scholar] [CrossRef] [Green Version]
- Fischbeck, A.; Leucht, K.; Frey-Wagner, I.; Bentz, S.; Pesch, T.; Kellermeier, S.; Krebs, M.; Fried, M.; Rogler, G.; Hausmann, M.; et al. Sphingomyelin induces cathepsin D-mediated apoptosis in intestinal epithelial cells and increases infammation in DSS colitis. Gut 2011, 60, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Braun, A.; Treede, I.; Gotthardt, D.; Tietje, A.; Zahn, A.; Ruhwald, R.; Schoenfeld, U.; Welsch, T.; Kienle, P.; Erben, G.; et al. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: A clue to pathogenesis. Inflamm. Bowel. Dis. 2009, 15, 1705–1720. [Google Scholar] [CrossRef]
- Copple, B.; Li, T. Pharmacology of bile acid receptors: Evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol. Res. 2016, 104, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Devlin, A.; Fischbach, M. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 2015, 11, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2015, 62, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020, 27, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics Reveals Metabolic Biomarkers of Crohn’s Disease. PLoS ONE 2009, 4, 6386–6396. [Google Scholar] [CrossRef] [PubMed]
- Hartman, K.G.; Bortner, J.D.; Falk, G.W.; Yu, J.; Martín, M.G.; Rustgi, A.K.; Lynch, J.P. Modeling inflammation and oxidative stress in gastrointestinal disease development using novel organotypic culture systems. Stem Cell Res. Ther. 2013, 4, S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beni, M.; Pellegrini-Giampietro, D.E.; Moroni, F. A new endogenous anxiolytic agent: L-pyroglutamic acid. Fund. Clin. Pharmacol. 1988, 2, 77–82. [Google Scholar] [CrossRef]
- Keshavarzian, A. Excessive production of reactive oxygen metabolites by infamed colon: Analysis by chemiluminescence probe. Gastroenterology 1992, 103, 177–185. [Google Scholar] [CrossRef]
- Simmonds, N. Chemiluminescence assay of mucosal reactive oxygen metabolites in infammatory bowel disease. Gastroenterology 1992, 103, 186–196. [Google Scholar] [CrossRef]
- Ponnusamy, K.; Choi, J.N.; Kim, J.; Lee, S.Y.; Lee, C.H. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J. Med. Microbiol. 2011, 60, 817–827. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.L.; Guo, T.K.; Wang, Y.H.; Gao, M.T.; Qin, H.; Wu, Y.J. Therapeutic effect of ginsenoside rd in rats with TNBS-induced recurrent ulcerative colitis. Arch Pharm. Res. 2012, 35, 1231–1239. [Google Scholar] [CrossRef]
- Chou, Y.J.; Kober, K.M.; Kuo, C.H.; Yeh, K.H.; Kuo, T.C.; Tseng, Y.J.; Miaskowski, C.; Liang, J.T.; Shun, S.C. A pilot study of metabolomic pathways associated with fatigue in survivors of colorectal cancer. Biol. Res. Nurs. 2020, 23, 42–49. [Google Scholar] [CrossRef]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.S.; Jung, S.; Han, J.S.; Hwang, G.S.; Kim, M.K. LC/MS-based polar metabolite profiling identified unique biomarker signatures for cervical cancerand cervical intraepithelial neoplasia using global and targeted metabolomics. Cancers 2019, 11, 511–531. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Aa, J.; Xu, J.; Sun, M.; Qian, S.; Cheng, L.; Yang, S.; Shi, R. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. J. Gastroen. Hepatol. 2011, 26, 1290–1297. [Google Scholar] [CrossRef]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef]
- Jiwon, P.; Yumi, S.; Tae, H. Urinary metabolites as biomarkers for diagnosis of breast cancer: A preliminary study. J. Breast Dis. 2019, 7, 44–51. [Google Scholar]
- Zhang, Q.; Li, X.; Yin, X.; Wang, H.; Fu, C.; Wang, H.; Li, K.; Li, Y.; Zhang, X.; Liang, H.; et al. Metabolomic profiling reveals serum L-pyroglutamic acid as a potential diagnostic biomarker for systemic lupus erythematosus. Rheumatology 2021, 60, 598–606. [Google Scholar] [CrossRef]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. γH2AX and cancer. Nat. Rev. Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef]
- Mauri, G.; Arena, S.; Siena, S.; Bardelli, A.; Sartore-Bianchi, A. The DNA damage response pathway as a land of therapeutic opportunities for colorectal cancer. Ann. Oncol. 2020, 31, 1135–1147. [Google Scholar] [CrossRef]
- Anthony, T.; Andre, N. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar]
- Bui, T.M.; Butin-Israeli, V.; Wiesolek, H.L.; Zhou, M.; Rehring, J.F.; Wiesmüller, L.; Wu, J.D.; Yang, G.Y.; Hanauer, S.B.; Sebag, J.A.; et al. Neutrophils alter DNA repair landscape to impact survival and shape distinct therapeutic phenotypes of colorectal cancer. Gastroenterology 2021, 161, 225–238. [Google Scholar] [CrossRef]
- Alina, J.; Fiona, P.; Elizabeth, H. Host-microbiota maladaptation in colorectal cancer. Nature 2020, 585, 509–517. [Google Scholar]
- Russo, M.; Crisafulli, G.; Sogari, A.; Reilly, N.M.; Arena, S.; Lamba, S.; Bartolini, A.; Amodio, V.; Magrì, A.; Novara, L.; et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 2019, 366, 1473–1480. [Google Scholar] [CrossRef]




| m/z | RSD of Retention Time (%) | RSD of Peak Area (%) |
|---|---|---|
| 496.1399 | 0.0088 | 7.8288 |
| 524.3700 | 0.0829 | 5.5716 |
| 100.0762 | 0.0698 | 4.4323 |
| Metabolites | m/z | Rt (min) | FC | p-Value | VIP | Metabolic Pathways | Enzymes Genes |
|---|---|---|---|---|---|---|---|
| 3-Furoic acid | 113.0181 | 2.0264 | 1.23 | 7.70 × 10−4 | 3.77 | ||
| Pyroglutamic acid | 130.0465 | 2.0868 | 1.27 | 8.09 × 10−3 | 3.40 | Glutathione metabolism, glutathione synthetase | QPCT, OPLAH |
| (S)-2-Methylmalate | 149.0444 | 12.8028 | 0.65 | 3.56 × 10−2 | 1.99 | Fatty acid metabolism, lipid metabolism | |
| 3-Hydroxyanthranilic acid | 154.0442 | 2.0607 | 12.09 | 1.93 × 10−4 | 2.77 | Tryptophan metabolism | KYNU, HAAO, CAT |
| Glycylvaline | 175.1094 | 2.1603 | 1.48 | 4.45 × 10−3 | 1.61 | ||
| Paraxanthine | 181.0705 | 2.0425 | 1.40 | 4.93 × 10−5 | 1.33 | Caffeine metabolism | NAT1, NAT2, XDH |
| Leucodopachrome | 196.0561 | 8.1400 | 2.74 | 1.10 × 10−6 | 7.26 | Tyrosine metabolism | |
| L-Tryptophan | 205.0974 | 5.7118 | 0.83 | 1.45 × 10−3 | 9.65 | Tryptophan metabolism | DDC, IDO1, TPH1 |
| 3-(Dimethylamino) propyl benzoate | 208.1290 | 4.8659 | 6.81 | 2.255 × 10−3 | 1.82 | ||
| Glycyl-Phenylalanine | 223.1056 | 5.2276 | 1.27 | 1.74 × 10−2 | 1.22 | ||
| Leucylleucine | 245.1841 | 7.8416 | 0.66 | 3.56 × 10−3 | 2.36 | ||
| Glutamylvaline | 247.1279 | 3.5562 | 0.37 | 2.39 × 10−12 | 3.35 | ||
| gamma-Glutamylleucine | 261.1455 | 6.2629 | 0.57 | 8.60 × 10−10 | 2.86 | ||
| Oleamide | 282.2768 | 25.5358 | 1.76 | 1.20 × 10−4 | 1.05 | Fatty acid metabolism, lipid metabolism | FAAH, PLA2G2A |
| L-Octanoylcarnitine | 288.2164 | 15.4359 | 0.69 | 2.51 × 10−2 | 1.68 | Mitochondrial beta-oxidation of short-chain saturated fatty acids | CROT |
| Methyl linolenate | 293.2493 | 42.8410 | 0.70 | 1.99 × 10−8 | 4.07 | ||
| 17-Hydroxylinolenic acid | 295.2280 | 36.7773 | 0.11 | 1.04 × 10−8 | 1.60 | Fatty acid metabolism, lipid metabolism | |
| 3-Dehydrosphinganine | 300.2854 | 25.5358 | 1.78 | 6.96 × 10−4 | 2.16 | Sphingolipid metabolism | GBGT1, PIGL, SPTLC1 |
| Phenylalanyl phenylalanine | 313.1578 | 10.2882 | 0.70 | 9.95 × 10−3 | 6.74 | ||
| Decanoylcarnitine | 316.2458 | 19.5269 | 0.65 | 3.45 × 10−2 | 2.21 | Fatty acid metabolism, lipid metabolism | |
| Phytosphingosine | 318.3016 | 21.1592 | 1.33 | 7.22 × 10−3 | 4.95 | Sphingolipid metabolism | GBGT1, PIGL, PIGQ |
| Glycocholic acid | 448.3052 | 23.5106 | 0.16 | 5.79 × 10−6 | 1.08 | Bile acid biosynthesis | BAAT, GLYAT, GLYATL3 |
| Glycochenodeoxycholate | 450.3251 | 25.9395 | 0.13 | 1.34 × 10−5 | 2.44 | Bile acid biosynthesis | BAAT, GLYAT, GLYATL3 |
| LysoPC(15:0/0:0) | 482.3287 | 31.1631 | 0.68 | 2.90 × 10−4 | 2.16 | Glycerophospholipid metabolism, lipid metabolism | LYPLA1, PLA2G15 |
| LysoPC(16:1(9Z)/0:0) | 494.3247 | 30.8185 | 0.49 | 7.76 × 10−4 | 1.28 | Glycerophospholipid metabolism, lipid metabolism | LYPLA1, PLA2G15 |
| LysoPE(20:0) | 516.3382 | 16.7751 | 0.76 | 3.98 × 10−3 | 2.81 | Fatty acid metabolism, lipid metabolism | ENPP2 |
| LysoPC(18:1(9Z)/0:0) | 522.3610 | 35.3163 | 0.30 | 4.35 × 10−6 | 5.12 | Fatty acid metabolism, lipid metabolism | LYPLA1, PLA2G15 |
| LysoPE(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/0:0) | 526.2905 | 31.5744 | 1.36 | 2.40 × 10−3 | 1.91 | Glycerophospholipid metabolism, lipid metabolism | ENPP2 |
| LysoPE(22:0/0:0) | 560.3676 | 17.1108 | 0.75 | 2.84 × 10−3 | 3.11 | Glycerophospholipid metabolism, lipid metabolism | ENPP2 |
| PC(18:1(9Z)/16:1(9Z)) | 780.5490 | 51.6295 | 1.48 | 1.33 × 10−2 | 4.85 | Phosphatidylcholine biosynthesis | LYPLA1, PLA2G15 |
| PC(18:3(6Z,9Z,12Z)/18:1(9Z)) | 782.5640 | 51.6295 | 1.23 | 3.16 × 10−2 | 2.69 | Phosphatidylcholine biosynthesis | LYPLA1,PLA2G15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhang, R.; Xin, M.; Xu, Y.; Liu, S.; Yu, B.; Zhang, B.; Liu, J. Discovery and Validation of Potential Serum Biomarkers with Pro-Inflammatory and DNA Damage Activities in Ulcerative Colitis: A Comprehensive Untargeted Metabolomic Study. Metabolites 2022, 12, 997. https://doi.org/10.3390/metabo12100997
Li M, Zhang R, Xin M, Xu Y, Liu S, Yu B, Zhang B, Liu J. Discovery and Validation of Potential Serum Biomarkers with Pro-Inflammatory and DNA Damage Activities in Ulcerative Colitis: A Comprehensive Untargeted Metabolomic Study. Metabolites. 2022; 12(10):997. https://doi.org/10.3390/metabo12100997
Chicago/Turabian StyleLi, Mingxiao, Rui Zhang, Mingjie Xin, Yi Xu, Shijia Liu, Boyang Yu, Boli Zhang, and Jihua Liu. 2022. "Discovery and Validation of Potential Serum Biomarkers with Pro-Inflammatory and DNA Damage Activities in Ulcerative Colitis: A Comprehensive Untargeted Metabolomic Study" Metabolites 12, no. 10: 997. https://doi.org/10.3390/metabo12100997