Systematics of Ditaxinae and Related Lineages within the Subfamily Acalyphoideae (Euphorbiaceae) Based on Molecular Phylogenetics
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Taxon Sampling and Outgroup Selection
2.2. DNA Extraction, Amplification and Sequencing
2.3. Phylogenetic Analyses
3. Results
4. Discussion
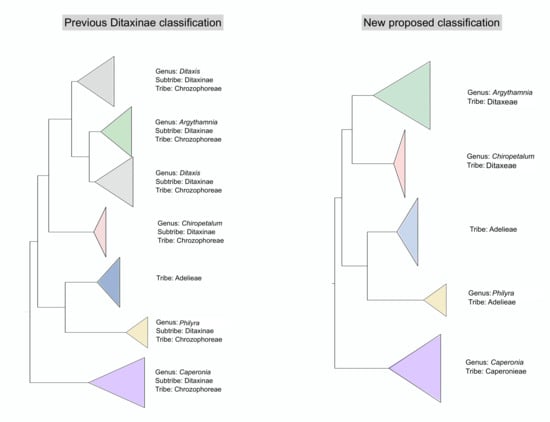
4.1. Changes in Generic Delimitation
4.2. Tribe Delimitation
4.3. Taxonomic Treatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chase, M.W.; Zmarzty, S.; Lledo, M.D.; Wurdack, K.J.; Swensen, S.M.; Fay, M.F. When in doubt, put it in Flacourtiaceae: A molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 2002, 57, 141–181. [Google Scholar] [CrossRef]
- Wurdack, K.J.; Hoffman, P.; Chase, M.W. Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto) using plastid rbcl and trnL-F DNA sequences. Am. J. Bot. 2005, 92, 1397–1420. [Google Scholar] [CrossRef]
- Tokuoka, T. Molecular phylogenetic analysis of Euphorbiaceae sensu stricto based on plastid and nuclear DNA sequences and ovule and seed character evolution. J. Plant Res. 2007, 120, 511–522. [Google Scholar] [CrossRef]
- Wurdack, K.J.; Davis, C.C. Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 2009, 96, 1551–1570. [Google Scholar] [CrossRef]
- Webster, G.L. Euphorbiaceae. In The Families and Genera of Vascular Plants. Flowering Plants. Eudicots. Malpighiales; Kubitzki, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 11, pp. 51–216. ISBN 978-3-642-39417-1. [Google Scholar]
- Wurdack, K.J.; Hoffmann, P.; Samuel, R.; Bruijn, A.; Bank, M.; Chase, M.W. Molecular phylogenetic analysis of Phyllanthaceae (Phyllanthoideae pro parte, Euphorbiaceae sensu lato) using plastid rbcL DNA sequences. Am. J. Bot. 2004, 91, 1882–1900. [Google Scholar] [CrossRef]
- Webster, G.L. Conspectus of a new classification of the Euphorbiaceae. Taxon 1975, 24, 593–601. [Google Scholar] [CrossRef] [Green Version]
- The Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 12385. [Google Scholar] [CrossRef] [Green Version]
- Govaerts, R.; Frodin, D.G.; Radcliffe-Smith, A. World Checklist and Bibliography of Euphorbiaceae (and Pandaceae); Royal Botanical Gardens, Kew: London, UK, 2000; Volume 2, p. 1661, ISBN-10: 1900347849. [Google Scholar]
- Radcliffe-Smith, A. Genera Euphorbiacearum; Royal Botanic Garden, Kew: London, UK, 2001; p. 464, ISBN-10: 1842460226. [Google Scholar]
- Pscheidt, A.C.; Cordeiro, I. Synopsis of the tribe Hippomaneae (Euphorbiaceae) in São Paulo State. Hoehnea 2012, 39, 347–368. [Google Scholar] [CrossRef]
- Grisebach, A.H.R. Flora of the Britrish West Indian Island; Rovell Reeve Co.: London, UK, 1859; Volume 2, pp. 193–315. [Google Scholar]
- Müller, A. Euphorbiaceae: Vorläufige Mitteilungen aus dem für DeCandolle Prodromus bestimmten Manuscript über diese Familie. Linnaea 1863, 34, 1–225. [Google Scholar]
- Pax, F.; Hoffmann, K. Euphorbiaceae—Acalypheae—Chrozophorinae. In Das Pflanzenreich, 4th ed.; Engler, A., Ed.; Engelmann: Leipzig, Germany, 1912; Volume 147, pp. 1–142. [Google Scholar]
- Külkamp, J. Chiropetalum A. Juss. (Euphorbiaceae Juss.) no Brasil. Master’s Thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil, 2018; p. 87. [Google Scholar]
- Külkamp, J.; Iganci, J.R.V.; Cordeiro, I.; Baumgratz, J.F.A. Ditaxis (Euphorbiaceae) from the Brazilian Caatinga, including a new species. Phytotaxa 2020, 455, 152–160. [Google Scholar] [CrossRef]
- Külkamp, J.; Caperonia in Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro. 2021. Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB17471 (accessed on 5 August 2021).
- Külkamp, J.; Ditaxis in Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro. 2021. Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB25551 (accessed on 5 August 2021).
- Külkamp, J.; Philyra in Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro. 2021. Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB25551 (accessed on 5 August 2021).
- Külkamp, J.; Iganci, J.R.V.; Cordeiro, I.; Chiropetalum in Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro 2021. Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB29160 (accessed on 4 August 2021).
- Külkamp, J.; Iganci, J.R.V.; Cordeiro, I.; Baumgratz, J.F. New species and occurrences of Caperonia (Euphorbiaceae) for South America. Phytotaxa 2021, 529, 86–92. [Google Scholar] [CrossRef]
- Ingram, J.W. A revisional study of Argythamnia subgenus Argythamnia (Euphorbiaceae). Gentes Herbarum 1967, 10, 1–38. [Google Scholar]
- Ingram, J.W. A revision of Argythamnia subgenus Chiropetalum (Euphorbiaceae). Gentes Herbarum 1980, 11, 437–468. [Google Scholar]
- Ingram, J.W. A sectional revision of Argythamnia subgenus Ditaxis (Euphorbiaceae). Ph.D. Thesis, University of California, Berkeley, CA, USA, 1956; p. 115. [Google Scholar]
- Ramírez-Amezcua, Y.; Steinmann, V.W. Revisión taxonómica de Argythamnia subgénero Ditaxis (Euphorbiaceae) en México. Bot. Sci. 2013, 91, 427–459. [Google Scholar] [CrossRef] [Green Version]
- Külkamp, J. Sistemática da Subtribo Ditaxinae (Euphorbiaceae). Ph.D. Thesis, Escola Nacional de Botânica Tropical, Jardim Botânico do Rio de Janeiro, Rio de Janeiro, Brazil, 2022; p. 245. [Google Scholar]
- Ingram, J.W. The generic limits of Argythamnia (Euphorbiaceae) defined. Gentes Herbarum 1980, 11, 427–436. [Google Scholar]
- Ramírez-Amezcua, Y. Relaciones Filogenéticas en Argythamnia (Euphorbiaceae) Sensu Lato. Master’s Thesis, Universidad Michoacana de San Nicolás de Hidalgo, Michoacán, Mexico, 2011; p. 64. [Google Scholar]
- Jestrow, B.; Gutièrez, J.A.; Francisco-Ortega, J. Islands within islands: A molecular phylogenetic study of the Leucocroton alliance (Euphorbiaceae) across the Carribean Islands and within the serpentinite archipelago of Cuba. J. Biogeogr. 2012, 39, 452–464. [Google Scholar] [CrossRef]
- Cervantes, A.; Fuentes, S.; Gutiérrez, J.; Magallón, S.; Borsch, T. Successive arrivals since the Miocene shaped the diversity of the Caribbean Acalyphoideae (Euphorbiaceae). J. Biogeogr. 2016, 43, 1773–1785. [Google Scholar] [CrossRef]
- Jestrow, B.; Rodríguez, F.J.; Francisco-Ortega, J. Generic delimitation in the Antillean Adelieae (Euphorbiaceae) with description of the Hispaniolan endemic genus Garciadelia. Taxon 2010, 59, 1801–1814. [Google Scholar] [CrossRef]
- Yang. Y.; Riina. R.; Morawetz, J.J.; Haevermans, T.; Aubriot, X.; Berry, P.E. Molecular phylogenetics and classification of Euphorbia subgenus Chamaesyce (Euphorbiaceae). Taxon 2012, 61, 764–789. [Google Scholar] [CrossRef] [Green Version]
- Riina, R.; Peirson, J.A.; Geltman, D.V.; Molero, J.; Frajman, B.; Pahlevani, A.; Barres, L.; Morawetz, J.J.; Salmaki, Y.; Zarre, S.; et al. A worldwide molecular phylogeny and classification of the leafy spurges, Euphorbia subgenus Esula (Euphorbiaceae). Taxon 2013, 62, 316–342. [Google Scholar] [CrossRef]
- Van Ee, B.W.; Riina, R.; Berry, P.E. A revised infrageneric classification and molecular phylogeny of New World Croton (Euphorbiaceae). Taxon 2011, 60, 791–823. [Google Scholar] [CrossRef] [Green Version]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff; New York Botanical Garden’s Virtual Herbarium: New York, NY, USA, 2022; Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 20 February 2022).
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. ISBN 978-3-642-83962-7. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum- likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Masa-Iranzo, I.; Sanmartín, I.; Caruzo, M.B.R.; Riina, R. Skipping the Dry Diagonal: Spatio-temporal evolution of Croton section Cleodora (Euphorbiaceae) in the Neotropics. Bot. J. Linn. Soc. 2021, 197, 61–84. [Google Scholar] [CrossRef]
- Ronquist, F.; Deans, A.R. Bayesian phylogenetics and its influence on insect systematics. Annu. Rev. Entomol. 2010, 55, 189–206. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- O’Donell, C.A.; Lourteig, A. Chrozophorae Argentinae. Lilloa 1942, 8, 37–81. [Google Scholar]
- Baillon, H.E. D’Observations Botaniques. Adansonia 1864, 4, 289. [Google Scholar]
- Jestrow, B.; Proctor, G.; Francisco-Ortega, J. Lasiocroton trelawniensis (Euphorbiaceae), a critically endangered species from the Cockpit Country of Jamaica, belongs to Bernardia (Euphorbiaceae). Bot. Rev. 2008, 74, 166–177. [Google Scholar] [CrossRef]
- De-Nova, J.A.; Sosa, V. Phylogeny and generic delimitation of Adelia (Euphorbiaceae) inferred from molecular and morphological data. Taxon 2007, 56, 1027–1036. [Google Scholar] [CrossRef]
- Bentham, G. Contribution towards a Flora of South America—Enumeration of Plants Collected by Mr. Schomburgk in British Guiana. Lond. J. Bot. 1843, 2, 42–52. [Google Scholar]
- Borhidi, A. Taxonomic revision of genus Leucocroton (Euphorbiaceae). Acta Bot. Hung. 1991, 36, 13–40. [Google Scholar]
- De-Nova, J.A.; Sosa, V.; Wurdack, K.J. Phylogenetic relationships and the description of a new species of Enriquebeltrania (Euphorbiaceae s.s.): An enigmatic genus endemic to Mexico. Syst. Bot. 2006, 31, 533–546. [Google Scholar] [CrossRef]


| ITS | ETS | trnL-F | trnT-L | petD | cpDNA Combined | nDNA Combined | All Markers Combined | |
|---|---|---|---|---|---|---|---|---|
| Number of terminals/species | 170/86 | 90/61 | 174/85 | 65/53 | 41/30 | 178/80 | 177/81 | 223/85 |
| Aligned sequence length | 750 | 394 | 1053 | 593 | 1195 | 2841 | 1144 | 3985 |
| Missing data (%) | 8.3 | 4.5 | 4.5 | 3.3 | 16.3 | 49.8 | 24.8 | 54.5 |
| Model | GTR+I+G | GTR+I+G | TVMIV+G | TPM+I+G | GTR+G | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Külkamp, J.; Riina, R.; Ramírez-Amezcua, Y.; Iganci, J.R.V.; Cordeiro, I.; González-Páramo, R.; Lara-Cabrera, S.I.; Baumgratz, J.F.A. Systematics of Ditaxinae and Related Lineages within the Subfamily Acalyphoideae (Euphorbiaceae) Based on Molecular Phylogenetics. Biology 2023, 12, 173. https://doi.org/10.3390/biology12020173
Külkamp J, Riina R, Ramírez-Amezcua Y, Iganci JRV, Cordeiro I, González-Páramo R, Lara-Cabrera SI, Baumgratz JFA. Systematics of Ditaxinae and Related Lineages within the Subfamily Acalyphoideae (Euphorbiaceae) Based on Molecular Phylogenetics. Biology. 2023; 12(2):173. https://doi.org/10.3390/biology12020173
Chicago/Turabian StyleKülkamp, Josimar, Ricarda Riina, Yocupitzia Ramírez-Amezcua, João R. V. Iganci, Inês Cordeiro, Raquel González-Páramo, Sabina Irene Lara-Cabrera, and José Fernando A. Baumgratz. 2023. "Systematics of Ditaxinae and Related Lineages within the Subfamily Acalyphoideae (Euphorbiaceae) Based on Molecular Phylogenetics" Biology 12, no. 2: 173. https://doi.org/10.3390/biology12020173