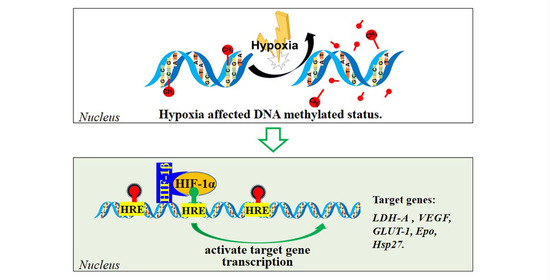
Hypoxia Affects HIF-1/LDH-A Signaling Pathway by Methylation Modification and Transcriptional Regulation in Japanese Flounder (Paralichthys olivaceus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Hypoxic Treatment and Sampling
2.2. Physiology, Biochemistry, and Hormone Measurements
2.2.1. Physiological Indicators in Blood
2.2.2. Biochemical Indicators in Serum
2.2.3. Hormonal Indicator in Serum
2.3. Genetic Structure and Phylogenetic Analysis of HIF-1α and LDH-A
2.4. Double In Situ Hybridization of HIF-1α and LDH-A Genes in Skeletal Muscle
2.5. Quantitative Real-Time PCR and Expression Analysis
2.6. Transcriptional Regulation Detection of HIF-1 on the LDH-A Gene
2.7. DNA Methylation Detection and Analysis
2.8. Statistical Analysis
3. Results
3.1. Effect of Acute Hypoxia on Physiology, Biochemistry, and Hormones
3.1.1. Blood Physiology (RBC, HGB, and WBC)
3.1.2. Blood Serum Biochemistry (LDH, ALP, ALT, GLU, TC, TG, and ALB)
Enzyme Activity in Blood Serum
Concentration in Blood Serum
3.1.3. COR Hormone
3.2. Genetic Structure and Phylogenetic Analysis of HIF-1α and LDH-A
3.3. The Colocalization and Expression of HIF-1α and LDH-A
3.4. HIF-1 Transcriptionally Regulates the LDH-A Gene
3.5. Methylation Modification and mRNA Expression of HIF-1α and LDH-A Genes
4. Discussion
4.1. Response to Hypoxia Stress
4.2. DNA Methylated Modification and Gene Expression
4.3. Transcriptional Regulation of HIF-1 and Its Target Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jenny, J.P.; Francus, P.; Normandeau, A.; Lapointe, F.; Perga, M.E.; Ojala, A.; Schimmelmann, A.; Zolitschka, B. Global spread of hypoxia in freshwater ecosystems during the last three centuries is caused by rising local human pressure. Glob. Change Biol. 2016, 22, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, M.; Storey, K.B. Metabolic response of longitudinal muscles to acute hypoxia in sea cucumber Apostichopus japonicus (Selenka): A metabolome integrated analysis. Comp. Biochem. Physiol. D-Genom. Proteom. 2019, 29, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Stramma, L.; Johnson, G.C.; Sprintall, J.; Mohrholz, V. Expanding Oxygen-Minimum Zones in the Tropical Oceans. Science 2008, 320, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Shezifi, Y.; Kimmel, E.; Diamant, A. The electrophysiological response of fish to hypoxia. Aquac. Eng. 1997, 16, 253–259. [Google Scholar] [CrossRef]
- Herbert, N.A.; Steffensen, J.F. The response of Atlantic cod, Gadus morhua, to progressive hypoxia: Fish swimming speed and physiological stress. Mar. Biol. 2005, 147, 1403–1412. [Google Scholar] [CrossRef]
- Tiedke, J.; Thiel, R.; Burmester, T. Molecular Response of Estuarine Fish to Hypoxia: A Comparative Study with Ruffe and Flounder from Field and Laboratory. PLoS ONE 2014, 9, e90778. [Google Scholar] [CrossRef]
- Hou, Z.S.; Wen, H.S.; Li, J.F.; He, F.; Li, Y.; Qi, X. Environmental hypoxia causes growth retardation, osteoclast differentiation and calcium dyshomeostasis in juvenile rainbow trout (Oncorhynchus mykiss). Sci. Total Environ. 2020, 705, 135272. [Google Scholar] [CrossRef]
- Greijer, A.; van der Groep, P.; Kemming, D.; Shvarts, A.; Semenza, G.; Meijer, G.; van de Wiel, M.; Belien, J.; van Diest, P.; van der Wall, E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 2005, 206, 291–304. [Google Scholar] [CrossRef]
- Hoppeler, H.; Vogt, M.; Weibel, E.R.; Flück, M. Response of skeletal muscle mitochondria to hypoxia. Exp. Physiol. 2003, 88, 109–119. [Google Scholar] [CrossRef]
- Hofer, T.; Wenger, R.H.; Gassmann, M. Oxygen sensing, HIF-1α stabilization and potential therapeutic strategies. Pflügers Arch.-Eur. J. Physiol. 2002, 443, 503–507. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1: Mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 2000, 88, 1474–1480. [Google Scholar] [CrossRef]
- Ke, Q.; Costa, M. Hypoxia-Inducible Factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef]
- Sharp, F.R.; Bernaudin, M. HIF1 and oxygen sensing in the brain. Nat. Rev. Neurosci. 2004, 5, 437–448. [Google Scholar] [CrossRef]
- Bruick, R.K.; McKnight, S.L. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef]
- Rundqvist, H.; Klintberg, M.; Norrbom, J.; Gustafsson, T.; Sundberg, C.J. HIF-1 specific prolyl hydroxylases in elite athletes. FASEB J. 2008, 22, 711–753.711. [Google Scholar] [CrossRef]
- Fan, S.; Li, X.; Lin, S.; Li, Y.; Ma, H.; Zhang, Z.; Qin, Z. Screening and Identification of Transcription Factors Potentially Regulating Foxl2 Expression in Chlamys farreri Ovary. Biology 2022, 11, 113. [Google Scholar] [CrossRef]
- Shi, Y.H.; Fang, W.G. Hypoxia-inducible factor-1 in tumour angiogenesis. World J. Gastroenterol. 2004, 10, 1082–1087. [Google Scholar] [CrossRef]
- Semenza, G.L.; Roth, P.H.; Fang, H.M.; Wang, G.L. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994, 269, 23757–23763. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A Gene Promoters Contain Essential Binding Sites for Hypoxia-inducible Factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Kaneda, M.; Sado, T.; Hata, K.; Okano, M.; Tsujimoto, N.; Li, E.; Sasaki, H. Role of De Novo DNA Methyltransferases in Initiation of Genomic Imprinting and X-Chromosome Inactivation. Cold Spring Harbor Symp. Quant. Biol. 2004, 69, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Beard, C.; Forster, A.C.; Bestor, T.H.; Jaenisch, R. DNA Methylation, Genomic Imprinting, and Mammalian Development. Cold Spring Harb. Symp. Quant. Biol. 1993, 58, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, R.P.; Patzel, K.A.; Martin, M.; Yasui, D.H.; Swanberg, S.E.; Hertz-Picciotto, I.; Hansen, R.L.; Water, J.V.d.; Pessah, I.N.; Jiang, R.; et al. MECP2 promoter methylation and X chromosome inactivation in autism. Autism Res. 2008, 1, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mehler, M.F. Epigenetic mechanisms underlying nervous system diseases. Handb. Clin. Neurol. 2018, 147, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Attwood, J.T.; Yung, R.L.; Richardson, B.C. DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci. 2002, 59, 241–257. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Lawe, D.; Ziff, E.B. Association of Myn, the Murine Homolog of Max, with c-Myc Stimulates Methylation-Sensitive DNA Binding and Ras Cotransformation. Cell 1991, 65, 395–407. [Google Scholar] [CrossRef]
- Uysal, F.; Ozturk, S.; Akkoyunlu, G. Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J. Assist. Reprod. Genet. 2018, 35, 503–513. [Google Scholar] [CrossRef]
- Bird, A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995, 11, 94–100. [Google Scholar] [CrossRef]
- Wu, S.; Huang, Y.; Li, S.; Wen, H.; Zhang, M.; Li, J.; Li, Y.; Shao, C.; He, F. DNA methylation levels and expression patterns of Smyd1a and Smyd1b genes during Metamorphosis of the Japanese Flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2018, 223, 16–22. [Google Scholar] [CrossRef]
- Huang, Y.; Wen, H.; Zhang, M.; Hu, N.; Si, Y.; Li, S.; He, F. The DNA methylation status of MyoD and IGF-I genes are correlated with muscle growth during different developmental stages of Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2018, 219–220, 33–43. [Google Scholar] [CrossRef]
- Huang, Y.; Hu, N.; Si, Y.; Li, S.; Wu, S.; Zhang, M.; Wen, H.; Li, J.; Li, Y.; He, F. Methylation status of the Follistatin gene at different development stages of Japanese Flounder (Paralichthys olivaceus). J. Ocean. Univ. China 2018, 17, 1243–1252. [Google Scholar] [CrossRef]
- Wan, J.; Oliver, V.F.; Wang, G.; Zhu, H.; Zack, D.J.; Merbs, S.L.; Qian, J. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genom. 2015, 16, 49. [Google Scholar] [CrossRef]
- Wang, S.; Pan, Z.; Zhang, Q.; Xie, Z.; Liu, H.; Li, Q. Differential mRNA expression and promoter methylation status of SYCP3 gene in testes of yaks and cattle-yaks. Reprod. Domest. Anim. 2012, 47, 455–462. [Google Scholar] [CrossRef]
- Sun, Y.; Li, R.; Zhai, G.; Zhang, X.; Wang, Y. DNA methylation of the PLIN1 promoter downregulates expression in chicken lines. Arch. Anim. Breed. 2019, 62, 375–382. [Google Scholar] [CrossRef]
- Vinoth, A.; Thirunalasundari, T.; Shanmugam, M.; Uthrakumar, A.; Suji, S.; Rajkumar, U. Evaluation of DNA methylation and mRNA expression of heat shock proteins in thermal manipulated chicken. Cell Stress Chaperones 2018, 23, 235–252. [Google Scholar] [CrossRef]
- Falisse, E.; Ducos, B.; Stockwell, P.A.; Morison, I.M.; Chatterjee, A.; Silvestre, F. DNA methylation and gene expression alterations in zebrafish early-life stages exposed to the antibacterial agent triclosan. Environ. Pollut. 2018, 243, 1867–1877. [Google Scholar] [CrossRef]
- Navarro-Martín, L.; Viñas, J.; Ribas, L.; Díaz, N.; Gutiérrez, A.; Croce, L.D.; Piferrer, F. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011, 7, e1002447. [Google Scholar] [CrossRef]
- Wang, M.; Qin, L.; Xie, C.; Li, W.; Yuan, J.; Kong, L.; Yu, W.; Xia, G.; Liu, S. Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant Cell Physiol. 2014, 55, 1354–1365. [Google Scholar] [CrossRef]
- Li, H.; Tong, L.; Tao, H.; Liu, Z. Genome-wide analysis of the hypoxia-related DNA methylation-driven genes in lung adenocarcinoma progression. Biosci. Rep. 2020, 40, 1–14. [Google Scholar] [CrossRef]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Gene. Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Morales, E.; Meier, K.; Sandoval-Carrillo, A.; Salas-Pacheco, J.; Vázquez-Cárdenas, P.; Arias-Carrión, O. Implications of DNA Methylation in Parkinson’s Disease. Front. Mol. Neurosci. 2017, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, C.; Li, X.; Wang, Z.; Wu, J.; Qian, Y.; Wei, Y. Associations between metal exposure and global DNA methylation in potentially affected people in E-Waste recycling sites in Taizhou City, China. Sci. Total Environ. 2020, 711, 135100. [Google Scholar] [CrossRef] [PubMed]
- Tidwell, J.H.; Allan, G.L. Fish as food: Aquaculture’s contribution. EMBO Rep. 2001, 2, 958–963. [Google Scholar] [CrossRef]
- Breen, E.; Tang, K.; Olfert, M.; Knapp, A.; Wagner, P. Skeletal muscle capillarity during hypoxia: VEGF and its activation. High Alt. Med. Biol. 2008, 9, 158–166. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, F.; Liu, S.; Zhao, H.; Lu, W.; Zhang, Q. Transcriptomic Analysis Reveals Functional Interaction of mRNA-lncRNA-miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology 2022, 11, 213. [Google Scholar] [CrossRef]
- Duan, Y.; Dong, X.; Zhang, X.; Miao, Z. Effects of dissolved oxygen concentration and stocking density on the growth, energy budget and body composition of juvenile Japanese flounder, Paralichthys olivaceus (Temminck et Schlegel). Aquac. Res. 2011, 42, 407–416. [Google Scholar] [CrossRef]
- Lundby, C.; Calbet, J.A.L.; Robach, P. The response of human skeletal muscle tissue to hypoxia. Cell. Mol. Life Sci. 2009, 66, 3615–3623. [Google Scholar] [CrossRef]
- Slivka, D.R. Skeletal muscle response to hypoxia. Acta Physiol. 2017, 220, 9–10. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, X.; Qin, J.; Zong, S. Acute ammonia toxicity and gill morphological changes of Japanese flounder Paralichthys olivaceus in normal versus supersaturated oxygen. Aquac. Res. 2013, 44, 1752–1759. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Kotaki, T.; Yamada, Y.; Ohta, H. Ontogenic changes in tolerance to hypoxia and energy metabolism of larval and juvenile Japanese flounder Paralichthys olivaceus. J. Exp. Mar. Biol. Ecol. 2007, 352, 42–49. [Google Scholar] [CrossRef]
- Ni, M.; Wen, H.; Li, J.; Chi, M.; Bu, Y.; Ren, Y.; Zhang, M.; Song, Z.; Ding, H. Effects of stocking density on mortality, growth and physiology of juvenile Amur sturgeon (Acipenser schrenckii). Aquac. Res. 2016, 47, 1596–1604. [Google Scholar] [CrossRef]
- Kanda, S.; Okubo, K.; Oka, Y. Differential regulation of the luteinizing hormone genes in teleosts and tetrapods due to their distinct genomic environments—Insights into gonadotropin beta subunit evolution. Gen. Comp. Endocrinol. 2011, 173, 253–258. [Google Scholar] [CrossRef]
- Li, Q.; Wen, H.; Li, Y.; Zhang, Z.; Wang, L.; Mao, X.; Li, J.; Qi, X. FOXO1A promotes neuropeptide FF transcription subsequently regulating the expression of feeding-related genes in spotted sea bass (Lateolabrax maculatus). Mol. Cell. Endocrinol. 2020, 517, 110871. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Zhao, L.L.; Wu, H.; Liu, Q.; Liao, L.; Luo, J.; Lian, W.Q.; Cui, C.; Jin, L.; Ma, J.D.; et al. Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci. Total Environ. 2020, 713, 135157. [Google Scholar] [CrossRef] [PubMed]
- Caradec, J.; Sirab, N.; Keumeugni, C.; Revaud, D.; Loric, S. Reply: 18S is an appropriate housekeeping gene for in vitro hypoxia experiments. Br. J. Cancer 2010, 103, 591–592. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, S.; Zhang, J.; Wen, H.; Zhang, M.; He, F. Methylation status and expression patterns of myomaker gene play important roles in postnatal development in the Japanese flounder (Paralichthys olivaceus). Gen. Comp. Endocrinol. 2019, 280, 104–114. [Google Scholar] [CrossRef]
- Buttari, B.; Profumo, E.; Riganò, R. Crosstalk between Red Blood Cells and the Immune System and Its Impact on Atherosclerosis. Biomed Res. Int. 2015, 2015, 1–8. [Google Scholar] [CrossRef]
- Qiang, J.; Yang, H.; Wang, H.; Kpundeh, M.D.; Xu, P. Interacting effects of water temperature and dietary protein level on hematological parameters in Nile tilapia juveniles, Oreochromis niloticus (L.) and mortality under Streptococcus iniae infection. Fish Shellfish. Immunol. 2013, 34, 8–16. [Google Scholar] [CrossRef]
- Yang, S.; Yan, T.; Wu, H.; Xiao, Q.; Fu, H.M.; Luo, J.; Zhou, J.; Zhao, L.L.; Wang, Y.; Yang, S.Y.; et al. Acute hypoxic stress: Effect on blood parameters, antioxidant enzymes, and expression of HIF-1alpha and GLUT-1 genes in largemouth bass (Micropterus salmoides). Fish Shellfish. Immunol. 2017, 67, 449–458. [Google Scholar] [CrossRef]
- van der Weele, C.M.; Jeffery, W.R. Cavefish cope with environmental hypoxia by developing more erythrocytes and overexpression of hypoxia inducible genes. Elife 2021, 10, e69109. [Google Scholar] [CrossRef]
- Chen, N.; Wu, M.; Tang, G.-P.; Wang, H.-J.; Huang, C.-X.; Wu, X.-J.; He, Y.; Zhang, B.; Huang, C.-H.; Liu, H.; et al. Effects of Acute Hypoxia and Reoxygenation on Physiological and Immune Responses and Redox Balance of Wuchang Bream (Megalobrama amblycephala Yih, 1955). Front. Physiol. 2017, 8, 375. [Google Scholar] [CrossRef]
- Abhijith, B.D.; Ramesh, M.; Poopal, R.K. Responses of metabolic and antioxidant enzymatic activities in gill, liver and plasma of Catla catla during methyl parathion exposure. J. Basic Appl. Zool. 2016, 77, 31–40. [Google Scholar] [CrossRef]
- Das, P.C.; Ayyappan, S.; Jena, J.K.; Das, B.K. Acute toxicity of ammonia and its sub-lethal effects on selected haematological and enzymatic parameters of mrigal, Cirrhinus mrigala (Hamilton). Aquac. Res. 2004, 35, 134–143. [Google Scholar] [CrossRef]
- Muusze, B.; Marcon, J.; van den Thillart, G.; Almeida-Val, V. Hypoxia tolerance of Amazon fish Respirometry and energy metabolism of the cichlid Astronotus Ocellatus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1998, 120, 151–156. [Google Scholar] [CrossRef]
- Marti, H.H.; Jung, H.H.; Pfeilschifter, J.; Bauer, C. Hypoxia and cobalt stimulate lactate dehydrogenase (LDH) activity in vascular smooth muscle cells. Pflüeg. Arch.-Eur. J. Physiol. 1994, 429, 216–222. [Google Scholar] [CrossRef]
- Das, S.K.; Varadhan, S.; Gupta, G.; Mukherjee, S.; Dhanya, L.; Rao, D.N.; Vasudevan, D.M. Time-dependent effects of ethanol on blood oxidative stress parameters and cytokines. Indian J. Biochem. Biophys. 2009, 46, 116–121. [Google Scholar] [CrossRef]
- Nath, B.; Szabo, G. Hypoxia and hypoxia inducible factors: Diverse roles in liver diseases. Hepatology 2012, 55, 622–633. [Google Scholar] [CrossRef]
- Savransky, V.; Reinke, C.; Jun, J.; Bevans-Fonti, S.; Nanayakkara, A.; Li, J.; Myers, A.C.; Torbenson, M.S.; Polotsky, V.Y. Chronic intermittent hypoxia and acetaminophen induce synergistic liver injury. Exp. Physiol. 2009, 94, 228–239. [Google Scholar] [CrossRef]
- Wan, C.; Yin, P.; Xu, X.; Liu, M.; He, S.; Song, S.; Liu, F.; Xu, J. Effect of simulated transport stress on the rat small intestine: A morphological and gene expression study. Res. Vet. Sci. 2014, 96, 355–364. [Google Scholar] [CrossRef]
- Enes, P.; Panserat, S.; Kaushik, S.; Oliva-Teles, A. Nutritional regulation of hepatic glucose metabolism in fish. Fish Physiol. Biochem. 2009, 35, 519–539. [Google Scholar] [CrossRef]
- Dickerson, S.S.; Kemeny, M.E. Acute Stressors and Cortisol Responses: A Theoretical Integration and Synthesis of Laboratory Research. Psychol. Bull. 2004, 130, 355–391. [Google Scholar] [CrossRef]
- Mommsen, T.P.; Vijayan, M.M.; Moon, T.W. Cortisol in teleost: Dynamics, mechanisms of action, and metabolic regulation. Rev. Fish Biol. Fish. 1999, 9, 211–268. [Google Scholar] [CrossRef]
- Pickering, A.D.; Pottinger, T.G. Stress responses and disease resistance in salmonid fish: Effects of chronic elevation of plasma cortisol. Fish Physiol. Biochem. 1989, 7, 253–258. [Google Scholar] [CrossRef]
- Fuzzen, M.L.M.; Alderman, S.L.; Bristow, E.N.; Bernier, N.J. Ontogeny of the corticotropin-releasing factor system in rainbow trout and differential effects of hypoxia on the endocrine and cellular stress responses during development. Gen. Comp. Endocrinol. 2011, 170, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Gao, Z.; He, Q.; Zhou, D.; Guo, Z.; Ye, J. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am. J. Physiol.-Endocrinol. Metab. 2009, 296, E333–E342. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, J.; Wang, S.; Zhang, D.; Zhao, Y.; Wang, R.; Zhao, J. DNA Methylation Regulates Gene Expression in Intracranial Aneurysms. World Neurosurg. 2017, 105, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Virani, S.; Colacino, J.A.; Kim, J.H.; Rozek, L.S. Cancer Epigenetics: A Brief Review. ILAR J. 2012, 53, 359–369. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef]
- Ordovás, J.M.; Smith, C.E. Epigenetics and cardiovascular disease. Nat. Rev. Cardiol. 2010, 7, 510–519. [Google Scholar] [CrossRef]
- Xu, R.; Li, S.; Guo, S.; Zhao, Q.; Abramson, M.J.; Li, S.; Guo, Y. Environmental temperature and human epigenetic modifications: A systematic review. Environ. Pollut. 2020, 259, 113840. [Google Scholar] [CrossRef]
- Alvarez, K.; Andrés, M.C.d.; Takahashi, A.; Oreffo, R.O.C. Effects of hypoxia on anabolic and catabolic gene expression and DNA methylation in OA chondrocytes. BMC Musculoskelet. Disord. 2014, 15, 431. [Google Scholar] [CrossRef]
- Goyal, D.; Goyal, R. Angiogenic Transformation in Human Brain Micro Endothelial Cells: Whole Genome DNA Methylation and Transcriptomic Analysis. Front. Physiol. 2019, 10, 1502. [Google Scholar] [CrossRef]
- Li, Z.; Zhuang, X.; Zeng, J.; Tzeng, C.-M. Integrated Analysis of DNA Methylation and mRNA Expression Profiles to Identify Key Genes in Severe Oligozoospermia. Front. Physiol. 2017, 8, 261. [Google Scholar] [CrossRef]
- Jowaed, A.; Schmitt, I.; Kaut, O.; Wüllner, U. Methylation Regulates Alpha-Synuclein Expression and Is Decreased in Parkinson’s Disease Patients’ Brains. J. Neurosci. 2010, 30, 6355–6359. [Google Scholar] [CrossRef]
- Si, Y.; Ding, Y.; He, F.; Wen, H.; Li, J.; Zhao, J.; Huang, Z. DNA methylation level of cyp19a1a and Foxl2 gene related to their expression patterns and reproduction traits during ovary development stages of Japanese flounder (Paralichthys olivaceus). Gene 2016, 575, 321–330. [Google Scholar] [CrossRef]
- Thomson, K.; Game, J.; Karouta, C.; Morgan, I.G.; Ashby, R. Correlation between small-scale methylation changes and gene expression during the development of myopia. FASEB J. 2022, 36, e22129. [Google Scholar] [CrossRef]
- Soitamo, A.J.; Råbergh, C.M.I.; Gassmann, M.; Sistonen, L.; Nikinmaa, M. Characterization of a hypoxia-inducible factor (HIF-1α) from rainbow trout. J. Biol. Chem. 2001, 276, 19699–19705. [Google Scholar] [CrossRef]
- Kuo, C.-W.; Tsai, M.-H.; Lin, T.-K.; Tiao, M.-M.; Wang, P.-W.; Chuang, J.-H.; Chen, S.-D.; Liou, C.-W. mtDNA as a Mediator for Expression of Hypoxia-Inducible Factor 1α and ROS in Hypoxic Neuroblastoma Cells. Int. J. Mol. Sci. 2017, 18, 1220. [Google Scholar] [CrossRef]
- Xie, H.; Valera, V.A.; Merino, M.J.; Amato, A.M.; Signoretti, S.; Linehan, W.M.; Sukhatme, V.P.; Seth, P. LDH-A Inhibition, a therapeutic strategy for treatment of Hereditary Leiomyomatosis and Renal Cell Cancer (HLRCC). Mol. Cancer Ther. 2009, 8, 626–635. [Google Scholar] [CrossRef]
- Richards, J.G. Chapter 10 Metabolic and Molecular Responses of Fish To Hypoxia. In Fish Physiology; Elsevier Inc.: Amsterdam, The Netherlands, 2009; Volume 27, p. 443. [Google Scholar]
- Ergorul, C.; Ray, A.; Huang, W.; Wang, D.Y.; Ben, Y.; Cantuti-Castelvetri, I.; Grosskreutz, C.L. Hypoxia Inducible Factor-1α (HIF-1α) and Some HIF-1 Target Genes are Elevated in Experimental Glaucoma. J. Mol. Neurosci. 2010, 42, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Baptista, R.B.; Souza-Castro, N.; Almeida-Val, V.M.F. Acute hypoxia up-regulates HIF-1α and VEGF mRNA levels in Amazon hypoxia-tolerant Oscar (Astronotus ocellatus). Fish Physiol. Biochem. 2016, 42, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Thomas, P. Molecular cloning, characterization and expression of two hypoxia-inducible factor alpha subunits, HIF-1α and HIF-2α, in a hypoxia-tolerant marine teleost, Atlantic croaker (Micropogonias undulatus). Gene 2007, 396, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wang, Y. Epigenetic regulators: Multifunctional proteins modulating hypoxia-inducible factor-alpha protein stability and activity. Cell Mol. Life Sci. 2018, 75, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K. Cellular signal transduction of the hypoxia response. J. Biochem. 2009, 146, 757–765. [Google Scholar] [CrossRef]


















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Wen, H.; Yang, J.; Li, X.; Li, G.; Zhang, J.; Wu, S.; Butts, I.A.; He, F. Hypoxia Affects HIF-1/LDH-A Signaling Pathway by Methylation Modification and Transcriptional Regulation in Japanese Flounder (Paralichthys olivaceus). Biology 2022, 11, 1233. https://doi.org/10.3390/biology11081233
Liu B, Wen H, Yang J, Li X, Li G, Zhang J, Wu S, Butts IA, He F. Hypoxia Affects HIF-1/LDH-A Signaling Pathway by Methylation Modification and Transcriptional Regulation in Japanese Flounder (Paralichthys olivaceus). Biology. 2022; 11(8):1233. https://doi.org/10.3390/biology11081233
Chicago/Turabian StyleLiu, Binghua, Haishen Wen, Jun Yang, Xiaohui Li, Guangling Li, Jingru Zhang, Shuxian Wu, Ian AE Butts, and Feng He. 2022. "Hypoxia Affects HIF-1/LDH-A Signaling Pathway by Methylation Modification and Transcriptional Regulation in Japanese Flounder (Paralichthys olivaceus)" Biology 11, no. 8: 1233. https://doi.org/10.3390/biology11081233