Inhibitors of ATP Synthase as New Antibacterial Candidates
Abstract
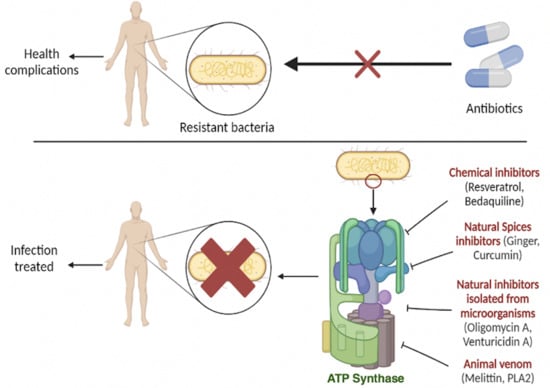
:1. Introduction
2. Structure and Function of ATP Synthase
3. Therapeutical Applications
3.1. Chemical Inhibitors
3.1.1. Resveratrol
3.1.2. Piceatannol
3.1.3. Bedaquiline
3.1.4. Tomatidine
3.1.5. N,N-dicyclohexylcarbodiimide
3.2. Inhibitors Isolated from Bacteria
3.2.1. Oligomycin A
3.2.2. Venturicidin A
3.3. Natural Inhibitors
3.3.1. Natural Spices
3.3.2. Animal Venoms
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capaldi, R.A.; Aggeler, R.; Turina, P.; Wilkens, S. Coupling between catalytic sites and the proton channel in F1F0-type ATPases. Trends Biochem. Sci. 1994, 19, 284–289. [Google Scholar] [CrossRef]
- Nijtmans, L.G.; Klement, P.; Houštěk, J.; van den Bogert, C. Assembly of mitochondrial ATP synthase in cultured human cells: Implications for mitochondrial diseases. Biochim. Biophys. Acta-Mol. Basis Dis. 1995, 1272, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Zeviani, M.; Di Donato, S. Mitochondrial disorders. Brain 2004, 127, 2153–2172. [Google Scholar] [CrossRef]
- Ahmad, Z.; Cox, J.L. ATP Synthase: The Right Size Base Model for Nanomotors in Nanomedicine. Sci. World J. 2014, 2014, 567398. [Google Scholar] [CrossRef] [Green Version]
- Devenish, R.J.; Prescott, M.; Rodgers, A.J. The Structure and Function of Mitochondrial F1F0–ATP Synthases. Int. Rev. Cell Mol. Biol. 2008, 267, 1–58. [Google Scholar] [CrossRef]
- Vik, S.B.; Long, J.C.; Wada, T.; Zhang, D. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim. Biophys. Acta-Bioenerg. 2000, 1458, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Noji, H.; Ueno, H.; McMillan, D.G.G. Catalytic robustness and torque generation of the F1-ATPase. Biophys. Rev. 2017, 9, 103–118. [Google Scholar] [CrossRef] [Green Version]
- Okuno, D.; Iino, R.; Noji, H. Rotation and structure of FoF1-ATP synthase. J. Biochem. 2011, 149, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi-Matsui, M.; Sekiya, M.; Futai, M. ATP synthase from Escherichia coli: Mechanism of rotational catalysis, and inhibition with the ε subunit and phytopolyphenols. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 129–140. [Google Scholar] [CrossRef]
- Guo, H.; Suzuki, T.; Rubinstein, J.L. Structure of a bacterial ATP synthase. eLife 2019, 8, e43128. [Google Scholar] [CrossRef]
- Iino, R.; Hasegawa, R.; Tabata, K.V.; Noji, H. Mechanism of inhibition by C-terminal alpha-helices of the epsilon subunit of Escherichia coli FoF1-ATP synthase. J. Biol. Chem. 2009, 284, 17457–17464. [Google Scholar] [CrossRef] [Green Version]
- Iino, R.; Murakami, T.; Iizuka, S.; Kato-Yamada, Y.; Suzuki, T.; Yoshida, M. Real-time monitoring of conformational dynamics of the epsilon subunit in F1-ATPase. J. Biol. Chem. 2005, 280, 40130–401304. [Google Scholar] [CrossRef] [Green Version]
- Feniouk, B.A.; Suzuki, T.; Yoshida, M. The role of subunit epsilon in the catalysis and regulation of FOF1-ATP synthase. Biochim. Biophys. Acta-Bioenerg. 2006, 1757, 326–338. [Google Scholar] [CrossRef] [Green Version]
- Fillingame, R.H.; Angevine, C.M.; Dmitriev, O.Y. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 2003, 555, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Handbook of ATPases; Wiley Online Books. Wiley Online Library, Weinheim, Germany. Available online: https://onlinelibrary.wiley.com/doi/book/10.1002/3527606122#page=254 (accessed on 16 November 2022).
- Jiang, W.; Hermolin, J.; Fillingame, R.H. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. USA 2001, 98, 4966–4971. [Google Scholar] [CrossRef] [Green Version]
- Mitome, N.; Suzuki, T.; Hayashi, S.; Yoshida, M. Thermophilic ATP synthase has a decamer c-ring: Indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc. Natl. Acad. Sci. USA 2004, 101, 12159–12164. [Google Scholar] [CrossRef] [Green Version]
- Meier, T.; Polzer, P.; Diederichs, K.; Welte, W.; Dimroth, P. Structure of the Rotor Ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science 2022, 308, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Stahlberg, H.; Müller, D.J.; Suda, K.; Fotiadis, D.; Engel, A.; Meier, T. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2001, 2, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Meier, T.; Ferguson, S.A.; Cook, G.M.; Dimroth, P.; Vonck, J. Structural Investigations of the Membrane-Embedded Rotor Ring of the F-ATPase from Clostridium paradoxum. J. Bacteriol. 2022, 188, 7759–7764. [Google Scholar] [CrossRef] [Green Version]
- Meier, T.; Morgner, N.; Matthies, D.; Pogoryelov, D.; Keis, S.; Cook, G.M.; Dimroth, P.; Brutschy, B. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol. Microbiol. 2007, 65, 1181–1192. [Google Scholar] [CrossRef]
- Preiss, L.; Yildiz, O.; Hicks, D.B.; Krulwich, T.A.; Meier, T. A new type of proton coordination in an F(1)F(o)-ATP synthase rotor ring. PLoS Biol. 2010, 8, e1000443. [Google Scholar] [CrossRef]
- Watt, I.N.; Montgomery, M.G.; Runswick, M.J.; Leslie, A.G.W.; Walker, J.E. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. USA 2010, 107, 16823–16827. [Google Scholar] [CrossRef] [Green Version]
- Neupane, P.; Bhuju, S.; Thapa, N.; Bhattarai, H.K. ATP Synthase: Structure, Function and Inhibition. Biomol. Concepts 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Diez, M.D.A.; Zimmermann, B.; Börsch, M.; König, M.; Schweinberger, E.; Steigmiller, S.; Reuter, R.; Felekyan, S.; Kudryavtsev, V.; Seidel, C.; et al. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat. Struct. Mol. Biol. 2004, 11, 135–141. [Google Scholar] [CrossRef]
- Symersky, J.; Osowski, D.; Walters, D.E.; Mueller, D.M. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA 2012, 109, 13961–13965. [Google Scholar] [CrossRef] [Green Version]
- Stock, D.; Leslie, A.G.W.; Walker, J.E. Molecular Architecture of the Rotary Motor in ATP Synthase. Science 1999, 286, 1700–1705. [Google Scholar] [CrossRef] [Green Version]
- Nøhr-Meldgaard, K.; Ovsepian, A.; Ingmer, H.; Vestergaard, M. Resveratrol enhances the efficacy of aminoglycosides against Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 390–396. [Google Scholar] [CrossRef]
- Ahmad, Z.; Ahmad, M.; Okafor, F.; Jones, J.; Abunameh, A.; Cheniya, R.P.; Kady, I.O. Effect of structural modulation of polyphenolic compounds on the inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2012, 50, 476–486. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Beck, C.; Nøhr-Meldgaard, K.; Peschel, A.; Kretschmer, D.; Ingmer, H.; Vestergaard, M. Inhibition of the ATP synthase sensitizes Staphylococcus aureus towards human antimicrobial peptides. Sci. Rep. 2020, 10, 11391. [Google Scholar] [CrossRef]
- Salehi, B.; Mishra, A.P.; Nigam, M.; Sener, B.; Kilic, M.; Sharifi--Rad, M.; Fokou, P.V.T.; Martins, N.; Sharifi-Rad, J. Resveratrol: A Double-Edged Sword in Health Benefits. Biomedicines 2018, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Tomé-Carneiro, J.; Gonzálvez, M.; Larrosa, M.; Yáñez-Gascón, M.J.; García-Almagro, F.J.; Ruiz-Ros, J.A.; Tomas-Barberan, F.; Conesa, M.T.G.; Espín, J.C. Grape Resveratrol Increases Serum Adiponectin and Downregulates Inflammatory Genes in Peripheral Blood Mononuclear Cells: A Triple-Blind, Placebo-Controlled, One-Year Clinical Trial in Patients with Stable Coronary Artery Disease. Cardiovasc. Drugs Ther. 2013, 27, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, M.; Nakamoto, R.K.; Nakanishi-Matsui, M.; Futai, M. Binding of Phytopolyphenol Piceatannol Disrupts β/γ Subunit Interactions and Rate-limiting Step of Steady-state Rotational Catalysis in Escherichia coli F1-ATPase. J. Biol. Chem. 2012, 287, 22771–22780. [Google Scholar] [CrossRef] [Green Version]
- Dadi, P.K.; Ahmad, M.; Ahmad, Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. Int. J. Biol. Macromol. 2009, 45, 72–79. [Google Scholar] [CrossRef]
- Sekiya, M.; Izumisawa, S.; Iwamoto-Kihara, A.; Fan, Y.; Shimoyama, Y.; Sasaki, M.; Nakanishi-Matsui, M. Proton-pumping F-ATPase plays an important role in Streptococcus mutans under acidic conditions. Arch. Biochem. Biophys. 2019, 666, 46–51. [Google Scholar] [CrossRef]
- Kershaw, J.; Kim, K.-H. The Therapeutic Potential of Piceatannol, a Natural Stilbene, in Metabolic Diseases: A Review. J. Med. Food 2017, 20, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.-L.; Chan, S.-W. A Review of the Pharmacological Effects of Piceatannol on Cardiovascular Diseases. Phytother. Res. 2014, 28, 1581–1588. [Google Scholar] [CrossRef]
- WHO|Regional Office for Africa. Tuberculosis (TB). Available online: https://www.afro.who.int/health-topics/tuberculosis-tb (accessed on 30 October 2022).
- Haagsma, A.C.; Abdillahi-Ibrahim, R.; Wagner, M.J.; Krab, K.; Vergauwen, K.; Guillemont, J.; Andries, K.; Lill, H.; Koul, A.; Bald, D. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob. Agents Chemother. 2009, 53, 1290–1292. [Google Scholar] [CrossRef] [Green Version]
- Koul, A.; Vranckx, L.; Dendouga, N.; Balemans, W.; Van den Wyngaert, I.; Vergauwen, K.; Göhlmann, H.W.; Willebrords, R.; Poncelet, A.; Guillemont, J.; et al. Diarylquinolines Are Bactericidal for Dormant Mycobacteria as a Result of Disturbed ATP Homeostasis. J. Biol. Chem. 2008, 283, 25273–25280. [Google Scholar] [CrossRef] [Green Version]
- MSF Southern Africa. MSF Demand J & J Reduce Price of anti-TB Drug. Available online: https://www.msf.org.za/news-and-resources/press-release/msf-demands-johnson-johnson-reduce-price-life-saving-tb-drug (accessed on 30 October 2022).
- World Health Organization. Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- World Health Organization. The Selection and Use of Essential Medicines; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Haagsma, A.C.; Podasca, I.; Koul, A.; Andries, K.; Guillemont, J.; Lill, H.; Bald, D. Probing the Interaction of the Diarylquinoline TMC207 with Its Target Mycobacterial ATP Synthase. PLoS ONE 2011, 6, e23575. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Courbon, G.M.; Bueler, S.A.; Mai, J.; Liu, J.; Rubinstein, J.L. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 2021, 589, 143–147. [Google Scholar] [CrossRef]
- Biuković, G.; Basak, S.; Manimekalai, M.S.S.; Rishikesan, S.; Roessle, M.; Dick, T. Variations of Subunit ε of the Mycobacterium tuberculosis F1Fo ATP Synthase and a Novel Model for Mechanism of Action of the Tuberculosis Drug TMC207. Antimicrob. Agents Chemother. 2013, 57, 168–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krah, A.; Grüber, G.; Bond, P.J. Binding properties of the anti-TB drugs bedaquiline and TBAJ-876 to a mycobacterial F-ATP synthase. Curr. Res. Struct. Biol. 2022, 4, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Goöhlmann, H.W.H.; Neefs, J.-M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A Diarylquinoline Drug Active on the ATP Synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Hotra, A.; Ragunathan, P.; Ng, P.S.; Seankongsuk, P.; Harikishore, A.; Sarathy, J.P.; Saw, W.; Lakshmanan, U.; Sae-Lao, P.; Kalia, N.P.; et al. Discovery of a Novel Mycobacterial F-ATP Synthase Inhibitor and its Potency in Combination with Diarylquinolines. Angew. Chem. Int. Ed. 2020, 59, 13295–13304. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Lamprecht, D.A.; Asmal, R.; Adamson, J.H.; Borah, K.; Beste, D.J.V.; Lee, B.S.; Pethe, K.; Rousseau, S.; Krieger, I.; et al. Bedaquiline reprograms central metabolism to reveal glycolytic vulnerability in Mycobacterium tuberculosis. Nat. Commun. 2020, 11, 6092. [Google Scholar] [CrossRef]
- Giraud-Gatineau, A.; Coya, J.M.; Maure, A.; Biton, A.; Thomson, M.; Bernard, E.M. The antibiotic bedaquiline activates host macrophage innate immune resistance to bacterial infection. elife 2020, 9, e55692. [Google Scholar] [CrossRef]
- Gaida, R.; Truter, I.; Peters, C.A. Adverse effects of bedaquiline in patients with extensively drug-resistant tuberculosis. S. Afr. J. Infect. Dis. 2020, 35, 6. [Google Scholar] [CrossRef]
- Luo, M.; Zhou, W.; Patel, H.; Srivastava, A.P.; Symersky, J.; Bonar, M.M.; Faraldo-Gómez, J.D.; Liao, M.; Mueller, D.M. Bedaquiline inhibits the yeast and human mitochondrial ATP synthases. Commun. Biol. 2020, 3, 452. [Google Scholar] [CrossRef]
- Chagnon, F.; Guay, I.; Bonin, M.-A.; Mitchell, G.; Bouarab, K.; Malouin, F.; Marsault, É. Unraveling the structure–activity relationship of tomatidine, a steroid alkaloid with unique antibiotic properties against persistent forms of Staphylococcus aureus. Eur. J. Med. Chem. 2014, 80, 605–620. [Google Scholar] [CrossRef]
- Lamontagne Boulet, M.; Isabelle, C.; Guay, I.; Brouillette, E.; Langlois, J.P.; Jacques, P.É. Tomatidine Is a Lead Antibiotic Molecule That Targets Staphylococcus aureus ATP Synthase Subunit C. Antimicrob. Agents Chemother. 2018, 62, e02197-17. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, G.; Gattuso, M.; Grondin, G.; Marsault, É.; Bouarab, K.; Malouin, F. Tomatidine Inhibits Replication of Staphylococcus aureus Small-Colony Variants in Cystic Fibrosis Airway Epithelial Cells. Antimicrob. Agents Chemother. 2011, 55, 1937–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, G.; Lafrance, M.; Boulanger, S.; Séguin, D.L.; Guay, I.; Gattuso, M.; Marsault, É.; Bouarab, K.; Malouin, F. Tomatidine acts in synergy with aminoglycoside antibiotics against multiresistant Staphylococcus aureus and prevents virulence gene expression. J. Antimicrob. Chemother. 2012, 67, 559–568. [Google Scholar] [CrossRef] [Green Version]
- Langlois, J.-P.; Millette, G.; Guay, I.; Dubé-Duquette, A.; Chamberland, S.; Jacques, P.; Rodrigue, S.; Bouarab, K.; Marsault, É.; Malouin, F. Bactericidal Activity of the Bacterial ATP Synthase Inhibitor Tomatidine and the Combination of Tomatidine and Aminoglycoside Against Persistent and Virulent Forms of Staphylococcus aureus. Front. Microbiol. 2020, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Soltani, R.; Fazeli, H.; Bahri Najafi, R.; Jelokhanian, A. Evaluation of the Synergistic Effect of Tomatidine with Several Antibiotics against Standard and Clinical Isolates of Staphylococcus aureus, Enterococcus faecalis, Pseudomonas aeruginosa and Escherichia coli. Iran J. Pharm. Res. 2017, 16, 290–296. [Google Scholar] [PubMed]
- Boulanger, S.; Mitchell, G.; Bouarab, K.; Marsault, É.; Cantin, A.; Frost, E.H.; Déziel, E.; Malouin, F. Bactericidal Effect of Tomatidine-Tobramycin Combination against Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa Is Enhanced by Interspecific Small-Molecule Interactions. Antimicrob. Agents Chemother. 2015, 59, 7458–7464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guay, I.; Boulanger, S.; Isabelle, C.; Brouillette, E.; Chagnon, F.; Bouarab, K.; Marsault, É.; Malouin, F. Tomatidine and analog FC04-100 possess bactericidal activities against Listeria, Bacillus and Staphylococcus spp. BMC Pharmacol. Toxicol. 2018, 19, 7. [Google Scholar] [CrossRef] [Green Version]
- Toei, M.; Noji, H. Single-molecule Analysis of F0F1-ATP Synthase Inhibited by N,N-Dicyclohexylcarbodiimide. J. Biol. Chem. 2013, 288, 25717–25726. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Fisher, R.P.; Rienhöfer-Schweer, A.; Hoffschulte, H.K. DCCD inhibits protein translocation into plasma membrane vesicles from Escherichia coli at two different steps. EMBO J. 1987, 6, 3855–3861. [Google Scholar] [CrossRef]
- Vestergaard, M.; Roshanak, S.; Ingmer, H. Targeting the ATP Synthase in Staphylococcus aureus Small Colony Variants, Streptococcus pyogenes and Pathogenic Fungi. Antibiotics 2021, 10, 376. [Google Scholar] [CrossRef]
- Zhou, W.; Faraldo-Gómez, J.D. Membrane plasticity facilitates recognition of the inhibitor oligomycin by the mitochondrial ATP synthase rotor. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 789–796. [Google Scholar] [CrossRef]
- Milgrom, Y.M.; Duncan, T.M. Complex effects of macrolide venturicidins on bacterial F-ATPases likely contribute to their action as antibiotic adjuvants. Sci. Rep. 2021, 11, 13631. [Google Scholar] [CrossRef]
- Zharova, T.V.; Kozlovsky, V.S.; Grivennikova, V.G. Interaction of Venturicidin and Fo·F1-ATPase/ATP Synthase of Tightly Coupled Subbacterial Particles of Paracoccus denitrificans in Energized Membranes. Biochemistry 2022, 87, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Zharova, T.V.; Vinogradov, A.D. Functional heterogeneity of Fo·F1H+-ATPase/synthase in coupled Paracoccus denitrificans plasma membranes. Biochim. Biophys. Acta-Bioenerg. 2017, 1858, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Yarlagadda, V.; Medina, R.; Wright, G.D. Venturicidin A, A Membrane-active Natural Product Inhibitor of ATP synthase Potentiates Aminoglycoside Antibiotics. Sci. Rep. 2020, 10, 8134. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, D.E.; Houghton, R.L. Studies on Energy-Linked Reactions: Modified Mitochondrial ATPase of Oligomycin-Resistant Mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 1974, 46, 157–167. [Google Scholar] [CrossRef]
- Perlin, D.S.; Latchney, L.R.; Senior, A.E. Inhibition of Escherichia coli H+-ATPase by venturicidin, oligomycin and ossamycin. Biochim. Biophys. Acta-Bioenerg. 1985, 807, 238–244. [Google Scholar] [CrossRef]
- Sekiya, M.; Chiba, E.; Satoh, M.; Yamakoshi, H.; Iwabuchi, Y.; Futai, M.; Nakanishi-Matsui, M. Strong inhibitory effects of curcumin and its demethoxy analog on Escherichia coli ATP synthase F1 sector. Int. J. Biol. Macromol. 2014, 70, 241–245. [Google Scholar] [CrossRef]
- Sekiya, M.; Hisasaka, R.; Iwamoto-Kihara, A.; Futai, M.; Nakanishi-Matsui, M. A unique mechanism of curcumin inhibition on F1 ATPase. Biochem. Biophys. Res. Commun. 2014, 452, 940–944. [Google Scholar] [CrossRef]
- Hughes, T.; Azim, S.; Ahmad, Z. Inhibition of Escherichia coli ATP synthase by dietary ginger phenolics. Int. J. Biol. Macromol. 2021, 182, 2130–2143. [Google Scholar] [CrossRef]
- Salaramoli, S.; Mehri, S.; Yarmohammadi, F.; Hashemy, S.I.; Hosseinzadeh, H. The effects of ginger and its constituents in the prevention of metabolic syndrome: A review. Iran J. Basic Med. Sci. 2022, 25, 664–674. [Google Scholar]
- Qin, S.; Huang, L.; Gong, J.; Shen, S.; Huang, J.; Ren, H.; Hu, H. Efficacy and safety of turmeric and curcumin in lowering blood lipid levels in patients with cardiovascular risk factors: A meta-analysis of randomized controlled trials. Nutr. J. 2017, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Nehme, H.; Ayde, H.; El Obeid, D.; Sabatier, J.M.; Fajloun, Z. Potential Inhibitory Effect of Apis mellifera’s Venom and of Its Two Main Components-Melittin and PLA2-on Escherichia coli F1F0-ATPase. Antibiotics 2020, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Pedersen, P.L. ATP Synthase and the Actions of Inhibitors Utilized to Study Its Roles in Human Health, Disease, and Other Scientific Areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, H.; Tauseef, M.; Ahmad, Z. A connection between antimicrobial properties of venom peptides and microbial ATP synthase. Int. J. Biol. Macromol. 2018, 119, 23–31. [Google Scholar] [CrossRef]
- Haktanir, I.; Masoura, M.; Mantzouridou, F.T.; Gkatzionis, K. Mechanism of antimicrobial activity of honeybee (Apis mellifera) venom on Gram-negative bacteria: Escherichia coli and Pseudomonas spp. AMB Express 2021, 11, 54. [Google Scholar] [CrossRef]
- Kfoury, M.; Mouawad, C.; Rifi, M.; Sadek, R.; Sabatier, J.M.; Nehme, H.; Fajloun, Z. Montivipera bornmuelleri Venom: Inhibitory Effect on Staphylococcus epidermidis and Escherichia coli F1F0-ATPases and Cytotoxicity on HCT116 Cancer Cell Lines. Science 2021, 3, 31. [Google Scholar] [CrossRef]
- Amini, A.; Raheem, S.; Steiner, A.; Deeba, F.; Ahmad, Z. Insect venom peptides as potent inhibitors of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2020, 150, 23–30. [Google Scholar] [CrossRef]
- Azim, S.; McDowell, D.; Cartagena, A.; Rodriguez, R.; Laughlin, T.F.; Ahmad, Z. Venom peptides cathelicidin and lycotoxin cause strong inhibition of Escherichia coli ATP synthase. Int. J. Biol. Macromol. 2016, 87, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Bordon, K.; de CF Cologna, C.T.; Fornari-Baldo, E.C.; Pinheiro-Júnior, E.L.; Cerni, F.A.; Amorim, F.G. From Animal Poisons and Venoms to Medicines: Achievements, Challenges and Perspectives in Drug Discovery. Front. Pharmacol. 2020, 11, 1132. [Google Scholar] [CrossRef]
- Ahmad, Z. Venom peptides as selective inhibitors of bacterial ATP synthase. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Sung, S.-H.; Kim, J.-W.; Han, J.-E.; Shin, B.-C.; Park, J.-K.; Lee, G. Animal Venom for Medical Usage in Pharmacopuncture in Korean Medicine: Current Status and Clinical Implication. Toxins 2021, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Hotra, A.; Suter, M.; Biuković, G.; Ragunathan, P.; Kundu, S.; Dick, T. Deletion of a unique loop in the mycobacterial F-ATP synthase γ subunit sheds light on its inhibitory role in ATP hydrolysis-driven H(+) pumping. FEBS J. 2016, 283, 1947–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppe, J.; Schairer, H.U.; Sebald, W. Identification of Amino-Acid Substitutions in the Proteolipid Subunit of the ATP Synthase from Dicyclohexylcarbodiimide-Resistant Mutants of Escherichia coli. Eur. J. Biochem. 1980, 112, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppe, J.; Sebald, W. The proton conducting F0-part of bacterial ATP synthases. Biochim. Biophys. Acta-Rev. Bioenerg. 1984, 768, 1–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripoll-Rozada, J.; García-Cazorla, Y.; Getino, M.; Machón, C.; Sanabria-Ríos, D.; de la Cruz, F.; Cabezón, E.; Arechaga, I. Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol. Microbiol. 2016, 100, 912–921. [Google Scholar] [CrossRef]




| Species | Number of Subunit c Copies |
|---|---|
| E. coli | 10 |
| Thermophilic Bacillus PS3 | 10 |
| I. tartaricus | 11 |
| C. paradoxum | 11 |
| Thermoalkaliphilic Bacillus TA2, TA1 | 13 |
| B. pseudofirmus OF4 | 13 |
| Bovine Mitochondria | 8 |
| ATP Synthase Inhibitor | Targeted Subunit | Bacteria | Reference |
|---|---|---|---|
| Resveratrol | subunit β and c-terminal region of subunit γ | S. aureus | [28] |
| Venturicidin A | subunit c | E. coli P. aeruginosa P. denitrificans MRSA Enterococcus | [66,67,68,69] |
Bedaquiline | subunit c and ε | M. tuberculosis Selective effect against: Nocardia Corynebacterium, S. pneumonia S. aureus E. faecalis E. coli, H. pylori, H. influenza | [39,40,41,42,43,48] |
| Tomatidine | subunit c | S. aureus P. aeruginosa E. faecalis L. monocytogenes | [54,56,57,59,61] |
| Piceatannol | Pocket created by α and β stator subunits and the carboxyl-terminal region of the subunit γ rotor | E. coli S. mutans | [34,35] |
| Oligomycin A DCCD | subunit c subunit c | ND E. coli | [65] [62,63] |
| Venoms Melittin LAAO PLA2 anoplin cupiennin 1a latarcin 1 latarcin 3a latarcin 5 pandinin 2 eumenitin lasiocepsin lycosin1 mastoparanB panurgine1 protonectin cathelicidin BF-30 lycotoxin | subunit β ND βDELSEED-motif βDELSEED-motif βDELSEED-motif βDELSEED-motif | E. coli Pseudomonas spp. S. epidermidis E. coli E. coli E. coli E. coli, P. aeruginosa S. aureus E. coli | [77,78,79,80] [81] [79] [82] [83] [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mackieh, R.; Al-Bakkar, N.; Kfoury, M.; Roufayel, R.; Sabatier, J.-M.; Fajloun, Z. Inhibitors of ATP Synthase as New Antibacterial Candidates. Antibiotics 2023, 12, 650. https://doi.org/10.3390/antibiotics12040650
Mackieh R, Al-Bakkar N, Kfoury M, Roufayel R, Sabatier J-M, Fajloun Z. Inhibitors of ATP Synthase as New Antibacterial Candidates. Antibiotics. 2023; 12(4):650. https://doi.org/10.3390/antibiotics12040650
Chicago/Turabian StyleMackieh, Rawan, Nadia Al-Bakkar, Milena Kfoury, Rabih Roufayel, Jean-Marc Sabatier, and Ziad Fajloun. 2023. "Inhibitors of ATP Synthase as New Antibacterial Candidates" Antibiotics 12, no. 4: 650. https://doi.org/10.3390/antibiotics12040650