Using Lactobacilli to Fight Escherichia coli and Staphylococcus aureus Biofilms on Urinary Tract Devices
Abstract
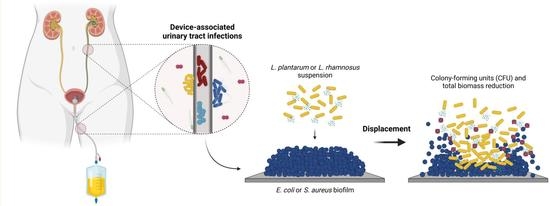
:1. Introduction
2. Results
2.1. Biofilm Cell Culturability
2.2. Biofilm Mass
3. Discussion
4. Materials and Methods
4.1. Preparation of Silicone Surfaces
4.2. Bacterial Strains and Culture Conditions
4.3. Influence of Probiotics on Pre-Formed Biofilms
4.3.1. Bacterial Enumeration
4.3.2. Biofilm Amount Determination
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention Catheter-associated Urinary Tract Infections (CAUTI)|HAI|CDC. Available online: https://www.cdc.gov/hai/ca_uti/uti.html (accessed on 20 September 2021).
- Siddiq, D.M.; Darouiche, R.O. New strategies to prevent catheter-associated urinary tract infections. Nat. Rev. Urol. 2012, 9, 305–314. [Google Scholar] [CrossRef]
- Maharjan, G.; Khadka, P.; Siddhi Shilpakar, G.; Chapagain, G.; Dhungana, G.R. Catheter-Associated Urinary Tract Infection and Obstinate Biofilm Producers. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 7624857. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization Report on the Burden of Endemic Health Care-Associated Infection Worldwide. 2011. Available online: https://apps.who.int/iris/handle/10665/80135 (accessed on 20 September 2021).
- Ramstedt, M.; Ribeiro, I.A.C.; Bujdakova, H.; Mergulhão, F.J.M.; Jordao, L.; Thomsen, P.; Alm, M.; Burmølle, M.; Vladkova, T.; Can, F.; et al. Evaluating efficacy of antimicrobial and antifouling materials for urinary tract medical devices: Challenges and recommendations. Macromol. Biosci. 2019, 19, e1800384. [Google Scholar] [CrossRef] [Green Version]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-associated infections, medical devices and biofilms: Risk, tolerance and control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.; Teixeira-Santos, R.; Gomes, L.C.; Faria, S.I.; Valcarcel, J.; Vázquez, J.A.; Cerqueira, M.A.; Pastrana, L.; Bourbon, A.I.; Mergulhão, F.J. Development of Chitosan-Based Surfaces to Prevent Single- and Dual-Species Biofilms of Staphylococcus aureus and Pseudomonas aeruginosa. Molecules 2021, 26, 4378. [Google Scholar] [CrossRef] [PubMed]
- Mandakhalikar, K.D.; Chua, R.R.; Tambyah, P.A. New Technologies for Prevention of Catheter Associated Urinary Tract Infection. Curr. Treat. Options Infect. Dis. 2016, 8, 24–41. [Google Scholar] [CrossRef]
- Tunney, M.M.; Gorman, S.P.; Patrick, S. Infection associated with medical devices. Int. J. Gen. Syst. 2002, 31, 195–205. [Google Scholar] [CrossRef]
- Vertes, A.; Hitchins, V.; Phillips, K.S. Analytical challenges of microbial biofilms on medical devices. Anal. Chem. 2012, 84, 3858–3866. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.S.; Almeida, C.; Melo, L.F.; Azevedo, N.F. Impact of polymicrobial biofilms in catheter-associated urinary tract infections. Crit. Rev. Microbiol. 2017, 43, 423–439. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef]
- Seif Eldein, S.S.; El-Temawy, A.-E.-K.A.; Ahmed, E.H. Biofilm Formation by E. coli Causing Catheter Associated Urinary Tract Infection ( CAUTI ) in Assiut University Hospital. Egypt. J. Med. Microbiol. 2013, 22, 101–110. [Google Scholar] [CrossRef]
- Niveditha, S.; Pramodhini, S.; Umadevi, S.; Kumar, S.; Stephen, S. The isolation and the biofilm formation of uropathogens in the patients with catheter associated urinary tract infections (UTIs). J. Clin. Diagn. Res. 2012, 6, 1478–1482. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, Z.; Wang, J.; Lopez, A.I.; Li, S.; Kumar, A.; Yu, F.; Chen, H.; Cai, C.; Zhang, L. Probiotic E. coli Nissle 1917 biofilms on silicone substrates for bacterial interference against pathogen colonization. Acta Biomater. 2017, 50, 353–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harb. Perspect. Biol. 2010, 2, a000398. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Schembri, M.A.; Klemm, P. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 2001, 69, 1322–1328. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, E.L.; Turner, I.G. Materials for urinary catheters: A review of their history and development in the UK. Med. Eng. Phys. 2005, 27, 443–453. [Google Scholar] [CrossRef]
- Stærk, K.; Grønnemose, R.B.; Palarasah, Y.; Kolmos, H.J.; Lund, L.; Alm, M.; Thomsen, P.; Andersen, T.E. A Novel Device-Integrated Drug Delivery System for Local Inhibition of Urinary Tract Infection. Front. Microbiol. 2021, 12, 1618. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Wang, Z.; Li, S.; Yuan, X. Antimicrobial strategies for urinary catheters. J. Biomed. Mater. Res.—Part A 2019, 107, 445–467. [Google Scholar] [CrossRef] [PubMed]
- Singha, P.; Locklin, J.; Handa, H. A review of the recent advances in antimicrobial coatings for urinary catheters. Acta Biomater. 2017, 50, 20–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AGN FAO; Nutrition and Consumer Protection Div; WHO, Geneva. FAO Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; FAO: Rome, Italy, 2006. [Google Scholar]
- Fioramonti, J.; Theodorou, V.; Bueno, L. Probiotics: What are they? What are their effects on gut physiology? Best Pract. Res. Clin. Gastroenterol. 2003, 17, 711–724. [Google Scholar] [CrossRef]
- Gogineni, V.K.; Morrow, L.E. Probiotics: Mechanisms of action and clinical applications. J. Probiotics Heal. 2013, 1, 101. [Google Scholar] [CrossRef] [Green Version]
- Aoudia, N.; Rieu, A.; Briandet, R.; Deschamps, J.; Chluba, J.; Jego, G.; Garrido, C.; Guzzo, J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Mosquera, A.; Alméciga-Díaz, C.J.; Melendez, A.P.; Sánchez, O.F. Fructooligosaccharides metabolism and effect on bacteriocin production in Lactobacillus strains isolated from ensiled corn and molasses. Anaerobe 2012, 18, 321–330. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Carr, F.J.; Chill, D.; Maida, N. The lactic acid bacteria: A literature survey. Crit. Rev. Microbiol. 2002, 28, 281–370. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Plaza-Díaz, J.; Muñoz-Quezada, S.; Gómez-Llorente, C.; Gil, A. Probiotic mechanisms of action. Ann. Nutr. Metab. 2012, 61, 160–174. [Google Scholar] [CrossRef]
- Ng, S.C.; Hart, A.L.; Kamm, M.A.; Stagg, A.J.; Knight, S.C. Mechanisms of action of probiotics: Recent advances. Inflamm. Bowel Dis. 2009, 15, 300–310. [Google Scholar] [CrossRef]
- Khalighi, A.; Behdani, R.; Kouhestani, S. Probiotics: A comprehensive review of their classification, mode of action and role in human nutrition. Probiotics Prebiotics Hum. Nutr. Health. 2016, 10, 63646. [Google Scholar]
- Ray Mohapatra, A.; Jeevaratnam, K. Inhibiting bacterial colonization on catheters: Antibacterial and antibiofilm activities of bacteriocins from Lactobacillus plantarum SJ33. J. Glob. Antimicrob. Resist. 2019, 19, 85–92. [Google Scholar] [CrossRef]
- Hasslöf, P.; Hedberg, M.; Twetman, S.; Stecksén-Blicks, C. Growth inhibition of oral mutans streptococci and candida by commercial probiotic lactobacilli—An in vitro study. BMC Oral Health 2010, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Vahedi Shahandashti, R.; Kasra Kermanshahi, R.; Ghadam, P. The inhibitory effect of bacteriocin produced by Lactobacillus acidophilus ATCC 4356 and Lactobacillus plantarum ATCC 8014 on planktonic cells and biofilms of Serratia marcescens. Turkish J. Med. Sci. 2016, 46, 1188–1196. [Google Scholar] [CrossRef]
- Jalilsood, T.; Baradaran, A.; Song, A.A.L.; Foo, H.L.; Mustafa, S.; Saad, W.Z.; Yusoff, K.; Rahim, R.A. Inhibition of pathogenic and spoilage bacteria by a novel biofilm-forming Lactobacillus isolate: A potential host for the expression of heterologous proteins. Microb. Cell Fact. 2015, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.M.; Teixeira-Santos, R.; Mergulhão, F.J.M.; Gomes, L.C. Effect of Lactobacillus plantarum Biofilms on the Adhesion of Escherichia coli to Urinary Tract Devices. Antibiotics 2021, 10, 966. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Feng, X.; Patel, R.; Patel, S.; Paranavitana, C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 2014, 14, 197. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Anti-biofilm properties of the fecal probiotic lactobacilli against Vibrio spp. Front. Cell. Infect. Microbiol. 2018, 8, 120. [Google Scholar] [CrossRef] [Green Version]
- Otero, M.C.; Nader-Macías, M.E. Inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim. Reprod. Sci. 2006, 96, 35–46. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Mahdi, S.; Khatibi, H.; Sharifi, S.; Memar, M.Y.; Vahed, S.Z. The battle of probiotics and their derivatives against biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.M.; Teixeira-Santos, R.; Mergulhão, F.J.M.; Gomes, L.C. The use of probiotics to fight biofilms in medical devices: A systematic review and meta-analysis. Microorganisms 2021, 9, 27. [Google Scholar] [CrossRef]
- Carvalho, F.M.; Teixeira-Santos, R.; Mergulhão, F.J.M.; Gomes, L.C. Targeting biofilms in medical devices using probiotic cells: A systematic review. AIMS Mater. Sci. 2021, 8, 501–523. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, D.H.; Song, K.Y.; Seo, K.H. Antimicrobial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J. Oral Microbiol. 2018, 10, 1472985. [Google Scholar] [CrossRef] [Green Version]
- Cadieux, P.; Watterson, J.D.; Denstedt, J.; Harbottle, R.R.; Puskas, J.; Howard, J.; Gan, B.S.; Reid, G. Potential application of polyisobutylene-polystyrene and a Lactobacillus protein to reduce the risk of device-associated urinary tract infections. Colloids Surf B Biointerfaces 2003, 28, 95–105. [Google Scholar] [CrossRef]
- Reid, G.; Tieszer, C. Use of lactobacilli to reduce the adhesion of Staphylococcus aureus to catheters. Int. Biodeterior. Biodegrad. 1994, 34, 73–83. [Google Scholar] [CrossRef]
- Gomes, L.C.; Silva, L.N.; Simões, M.; Melo, L.F.; Mergulhão, F.J. Escherichia coli adhesion, biofilm development and antibiotic susceptibility on biomedical materials. J. Biomed. Mater. Res. Part A 2015, 103, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, A.S.; Almeida, C.; Gomes, L.C.; Ferreira, C.; Mergulhão, F.J.; Melo, L.F.; Azevedo, N.F. An in vitro model of catheter-associated urinary tract infections to investigate the role of uncommon bacteria on the Escherichia coli microbial consortium. Biochem. Eng. J. 2017, 118, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Leonhard, M.; Moser, D.; Ma, S.; Schneider-Stickler, B. Inhibitory effect of probiotic lactobacilli supernatants on single and mixed non-albicans Candida species biofilm. Arch. Oral Biol. 2018, 85, 40–45. [Google Scholar] [CrossRef]
- Matsubara, V.H.; Wang, Y.; Bandara, H.M.H.N.; Mayer, M.P.A.; Samaranayake, L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016, 100, 6415–6426. [Google Scholar] [CrossRef] [Green Version]
- Rossoni, R.D.; de Barros, P.P.; de Alvarenga, J.A.; de Camargo Ribeiro, F.; dos Santos Velloso, M.; Fuchs, B.B.; Mylonakis, E.; Jorge, A.O.C.; Junqueira, J.C. Antifungal activity of clinical Lactobacillus strains against Candida albicans biofilms: Identification of potential probiotic candidates to prevent oral candidiasis. Biofouling 2018, 34, 212–225. [Google Scholar] [CrossRef] [Green Version]
- Fernández Ramírez, M.D.; Smid, E.J.; Abee, T.; Nierop Groot, M.N. Characterisation of biofilms formed by Lactobacillus plantarum WCFS1 and food spoilage isolates. Int. J. Food Microbiol. 2015, 207, 23–29. [Google Scholar] [CrossRef]
- Jaffar, N.; Ishikawa, Y.; Mizuno, K.; Okinaga, T.; Maeda, T. Mature biofilm degradation by potential probiotics: Aggregatibacter actinomycetemcomitans versus Lactobacillus spp. PLoS ONE 2016, 11, e0159466. [Google Scholar] [CrossRef]
- Song, Y.G.; Lee, S.H. Inhibitory effects of Lactobacillus rhamnosus and Lactobacillus casei on Candida biofilm of denture surface. Arch. Oral Biol. 2017, 76, 1–6. [Google Scholar] [CrossRef]
- Fayol-Messaoudi, D.; Berger, C.N.; Coconnier-Polter, M.H.; Liévin-Le Moal, V.; Servin, A.L. pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar typhimurium. Appl. Environ. Microbiol. 2005, 71, 6008–6013. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Chen, X.; Tu, Y.; Wang, S.; Chen, H. Effect of probiotic lactobacilli on the growth of Streptococcus mutans and multispecies biofilms isolated from children with active caries. Med. Sci. Monit. 2017, 23, 4175–4181. [Google Scholar] [CrossRef] [Green Version]
- Maldonado-Barragán, A.; Caballero-Guerrero, B.; Lucena-Padrós, H.; Ruiz-Barba, J.L. Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 2013, 33, 40–47. [Google Scholar] [CrossRef]
- Klaenhammer, T.R. Bacteriocins of lactic acid bacteria. Biochimie 1988, 70, 337–349. [Google Scholar] [CrossRef]
- McMillan, A.; Dell, M.; Zellar, M.P.; Cribby, S.; Martz, S.; Hong, E.; Fu, J.; Abbas, A.; Dang, T.; Miller, W.; et al. Disruption of urogenital biofilms by lactobacilli. Colloids Surf. B Biointerfaces 2011, 86, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Cadieux, P.A.; Burton, J.P.; Devillard, E.; Reid, G. Lactobacillus by-products inhibit the growth and virulence of uropathogenic Escherichia coli. J. Physiol. Pharmacol. 2009, 60, 13–18. [Google Scholar] [PubMed]
- Morais, I.M.C.; Cordeiro, A.L.; Teixeira, G.S.; Domingues, V.S.; Nardi, R.M.D.; Monteiro, A.S.; Alves, R.J.; Siqueira, E.P.; Santos, V.L. Biological and physicochemical properties of biosurfactants produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65. Microb. Cell Fact. 2017, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Ceresa, C.; Tessarolo, F.; Caola, I.; Nollo, G.; Cavallo, M.; Rinaldi, M.; Fracchia, L. Inhibition of Candida albicans adhesion on medical-grade silicone by a Lactobacillus-derived biosurfactant. J. Appl. Microbiol. 2015, 118, 1116–1125. [Google Scholar] [CrossRef]
- Sharma, D.; Saharan, B.S. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Reports 2016, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Zhang, J.; Qu, J.; Liu, J.; Yin, P.; Zhang, G.; Shang, D. Lactobacillus rhamnosus GG microcapsules inhibit Escherichia coli biofilm formation in coculture. Biotechnol. Lett. 2019, 41, 1007–1014. [Google Scholar] [CrossRef]
- Ahn, K.B.; Baik, J.E.; Park, O.J.; Yun, C.H.; Han, S.H. Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS ONE 2018, 13, e0192694. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.R.; Ahn, K.B.; Yun, C.H.; Park, O.J.; Perinpanayagam, H.; Yoo, Y.J.; Kum, K.Y.; Han, S.H. Lactobacillus plantarum lipoteichoic acid inhibits oral multispecies biofilm. J. Endod. 2019, 45, 310–315. [Google Scholar] [CrossRef]
- Teanpaisan, R.; Piwat, S.; Dahlén, G. Inhibitory effect of oral Lactobacillus against oral pathogens. Lett. Appl. Microbiol. 2011, 53, 452–459. [Google Scholar] [CrossRef]
- Alexandre, Y.; Le Berre, R.; Barbier, G.; Le Blay, G. Screening of Lactobacillus spp. for the prevention of Pseudomonas aeruginosa pulmonary infections. BMC Microbiol. 2014, 14, 107. [Google Scholar] [CrossRef] [Green Version]
- Todorov, S.; Gotcheva, B.; Dousset, X.; Onno, B.; Ivanova, I. Influence of growth medium on bacteriocin production in Lactobacillus plantarum ST31. Biotechnol. Biotechnol. Equip. 2000, 14, 50–55. [Google Scholar] [CrossRef]
- Brooks, T.; Keevil, C.W. A simple artificial urine for the growth of urinary pathogens. Lett. Appl. Microbiol. 1997, 24, 203–206. [Google Scholar] [CrossRef]
- Rodrigues, L.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Fracchia, L.; Cavallo, M.; Giovanna, M.; Banat, I.M. Biosurfactants and Bioemulsifiers Biomedical and Related Applications—Present Status and Future Potentials. In Biomedical Science, Engineering and Technology; Ghista, D.N., Ed.; InTech Publisher: London, UK, 2012; pp. 325–370. ISBN 978-953-307-471-9. [Google Scholar]
- Cerqueira, L.; Oliveira, J.A.; Nicolau, A.; Azevedo, N.F.; Vieira, M.J. Biofilm formation with mixed cultures of Pseudomonas aeruginosa/Escherichia coli on silicone using artificial urine to mimic urinary catheters. Biofouling 2013, 29, 829–840. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Growth of the Bacteriocin-Producing Lactobacillus sakei Strain CTC 494 in MRS Broth is Strongly Reduced Due to Nutrient Exhaustion: A Nutrient Depletion Model for the Growth of Lactic Acid Bacteria. Appl. Environ. Microbiol. 2001, 67, 4407–4413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, M.; Gomes, L.C.; Teixeira-Santos, R.; Pereira, M.F.R.; Soares, O.S.G.P.; Mergulhão, F.J. Optimizing CNT Loading in Antimicrobial Composites for Urinary Tract Application. Appl. Sci. 2021, 11, 4038. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, F.M.; Mergulhão, F.J.M.; Gomes, L.C. Using Lactobacilli to Fight Escherichia coli and Staphylococcus aureus Biofilms on Urinary Tract Devices. Antibiotics 2021, 10, 1525. https://doi.org/10.3390/antibiotics10121525
Carvalho FM, Mergulhão FJM, Gomes LC. Using Lactobacilli to Fight Escherichia coli and Staphylococcus aureus Biofilms on Urinary Tract Devices. Antibiotics. 2021; 10(12):1525. https://doi.org/10.3390/antibiotics10121525
Chicago/Turabian StyleCarvalho, Fábio M., Filipe J. M. Mergulhão, and Luciana C. Gomes. 2021. "Using Lactobacilli to Fight Escherichia coli and Staphylococcus aureus Biofilms on Urinary Tract Devices" Antibiotics 10, no. 12: 1525. https://doi.org/10.3390/antibiotics10121525