Detection of Sialic Acid to Differentiate Cervical Cancer Cell Lines Using a Sambucus nigra Lectin Biosensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Assembly of SNA Biosensors
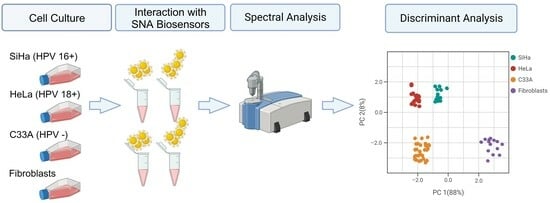
2.3. Cell lines and Culture
2.4. Cell Interaction with SNA Biosensors
2.5. ATR-FTIR Spectroscopy Spectra
2.6. Data Analysis
3. Results and Discussion
3.1. Characterization of SNA Biosensors
3.2. Infrared Spectroscopy
3.3. Discriminant Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirghani, H.; Jung, A.C.; Fakhry, C. Primary, Secondary and Tertiary Prevention of Human Papillomavirus-Driven Head and Neck Cancers. Eur. J. Cancer 2017, 78, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Purandare, N.C.; Patel, I.I.; Trevisan, J.; Bolger, N.; Kelehan, R.; von Bünau, G.; Martin-Hirsch, P.L.; Prendiville, W.J.; Martin, F.L. Biospectroscopy Insights into the Multi-Stage Process of Cervical Cancer Development: Probing for Spectral Biomarkers in Cytology to Distinguish Grades. Analyst 2013, 138, 3909. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical Cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Torres-Poveda, K.; Piña-Sánchez, P.; Vallejo-Ruiz, V.; Lizano, M.; Cruz-Valdez, A.; Juárez-Sánchez, P.; de la Garza-Salazar, J.; Manzo-Merino, J. Molecular Markers for the Diagnosis of High-Risk Human Papillomavirus Infection and Triage of Human Papillomavirus-Positive Women. Rev. Investig. Clin. 2020, 72, 198–212. [Google Scholar] [CrossRef]
- Piña-Sánchez, P. Human Papillomavirus: Challenges and Opportunities for the Control of Cervical Cancer. Arch. Med. Res. 2022, 53, 753–769. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, K.; Ahmadzai, A.A.; Valasoulis, G.; Trevisan, J.; Founta, C.; Nasioutziki, M.; Loufopoulos, A.; Kyrgiou, M.; Stasinou, S.M.; Karakitsos, P.; et al. Histology Verification Demonstrates That Biospectroscopy Analysis of Cervical Cytology Identifies Underlying Disease More Accurately than Conventional Screening: Removing the Confounder of Discordance. PLoS ONE 2014, 9, e82416. [Google Scholar] [CrossRef]
- Duraipandian, S.; Traynor, D.; Kearney, P.; Martin, C.; O’Leary, J.J.; Lyng, F.M. Raman Spectroscopic Detection of High-Grade Cervical Cytology: Using Morphologically Normal Appearing Cells. Sci. Rep. 2018, 8, 15048. [Google Scholar] [CrossRef] [PubMed]
- Pereira Viana, M.R.; Martins Alves Melo, I.; Pupin, B.; Raniero, L.J.; de Azevedo Canevari, R. Molecular Detection of HPV and FT-IR Spectroscopy Analysis in Women with Normal Cervical Cytology. Photodiagnosis Photodyn. Ther. 2020, 29, 101592. [Google Scholar] [CrossRef]
- Martinez-Morales, P.; Morán Cruz, I.; Roa-de la Cruz, L.; Maycotte, P.; Reyes Salinas, J.S.; Vazquez Zamora, V.J.; Gutierrez Quiroz, C.T.; Montiel-Jarquin, A.J.; Vallejo-Ruiz, V. Hallmarks of Glycogene Expression and Glycosylation Pathways in Squamous and Adenocarcinoma Cervical Cancer. PeerJ 2021, 9, e12081. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- López-Morales, D.; Reyes-Leyva, J.; Santos-López, G.; Zenteno, E.; Vallejo-Ruiz, V. Increased Expression of Sialic Acid in Cervical Biopsies with Squamous Intraepithelial Lesions. Diagn. Pathol. 2010, 5, 74. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, F.; Real, F.X. Differential Distribution of Sialic Acid in 2,3 and 2,6 Linkages in the Apical Membrane of Cultured Epithelial Cells and Tissues. J. Histochem. Cytochem. 2001, 49, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Domínguez, M.; Morales-Luna, G.; Mahlknecht, J.; Cheng, Q.; Aguilar-Hernández, I.; Ornelas-Soto, N. Optical Biosensors and Their Applications for the Detection of Water Pollutants. Biosensors 2023, 13, 370. [Google Scholar] [CrossRef] [PubMed]
- Vigneshvar, S.; Sudhakumari, C.C.; Senthilkumaran, B.; Prakash, H. Recent Advances in Biosensor Technology for Potential Applications—An Overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P. Biosensors and Their Applications—A Review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Mittal, S.; Das, M.; Saharia, A.; Tiwari, M. Optical Biosensors: A Decade in Review. Alexandria Eng. J. 2023, 67, 673–691. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical Biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef]
- Tai, J.; Fan, S.; Ding, S.; Ren, L. Gold Nanoparticles Based Optical Biosensors for Cancer Biomarker Proteins: A Review of the Current Practices. Front. Bioeng. Biotechnol. 2022, 10, 877193. [Google Scholar] [CrossRef]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-Free Optical Biosensors for Food and Biological Sensor Applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Ortega-Hernández, N.; Ortega-Romero, M.; Medeiros-Domingo, M.; Barbier, O.C.; Rojas-López, M. Detection of Biomarkers Associated with Acute Kidney Injury by a Gold Nanoparticle Based Colloidal Nano-Immunosensor by Fourier-Transform Infrared Spectroscopy with Principal Component Analysis. Anal. Lett. 2022, 55, 2370–2381. [Google Scholar] [CrossRef]
- Yang, X.; Sun, Z.; Low, T.; Hu, H.; Guo, X.; García de Abajo, F.J.; Avouris, P.; Dai, Q. Nanomaterial-Based Plasmon-Enhanced Infrared Spectroscopy. Adv. Mater. 2018, 30, 1704896. [Google Scholar] [CrossRef]
- López-Lorente, Á.I.; Mizaikoff, B. Recent Advances on the Characterization of Nanoparticles Using Infrared Spectroscopy. TrAC Trends Anal. Chem. 2016, 84, 97–106. [Google Scholar] [CrossRef]
- Zamudio Cañas, R.; Zaca Moran, O.; Jaramillo Flores, M.E.; Vallejo Ruiz, V.; Reyes Leyva, J.; Delgado Macuil, R.; Lopez Gayou, V. Characterization and Differentiation of Cervical Cancer Cell Lines Using ATR-FTIR Spectroscopy and Multivariate Data Analysis. Biomed. Signal Process. Control 2023, 86, 105169. [Google Scholar] [CrossRef]
- Sabbatini, S.; Conti, C.; Orilisi, G.; Giorgini, E. Infrared Spectroscopy as a New Tool for Studying Single Living Cells: Is There a Niche? Biomed. Spectrosc. Imaging 2017, 6, 85–99. [Google Scholar] [CrossRef]
- Su, K.-Y.; Lee, W.-L. Fourier Transform Infrared Spectroscopy as a Cancer Screening and Diagnostic Tool: A Review and Prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.L.S.; Gutiérrez, E.; Rodríguez, J.A.; Gomes, C.; David, L. Construction and Validation of a Sambucus Nigra Biosensor for Cancer-Associated STn Antigen. Biosens. Bioelectron. 2014, 57, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Avila, L.; Castro-Amaya, A.M.; Molina-Pineda, A.; Hernández-Gutiérrez, R.; Jave-Suarez, L.F.; Aguilar-Lemarroy, A. The Value of CXCL1, CXCL2, CXCL3, and CXCL8 as Potential Prognosis Markers in Cervical Cancer: Evidence of E6/E7 from HPV16 and 18 in Chemokines Regulation. Biomedicines 2023, 11, 2655. [Google Scholar] [CrossRef]
- Jagadeeshaprasad, M.G.; Govindappa, P.K.; Nelson, A.M.; Elfar, J.C. Isolation, Culture, and Characterization of Primary Schwann Cells, Keratinocytes, and Fibroblasts from Human Foreskin. J. Vis. Exp. 2022, 181, e63776. [Google Scholar] [CrossRef]
- Amina, S.J.; Guo, B. A Review on the Synthesis and Functionalization of Gold Nanoparticles as a Drug Delivery Vehicle. Int. J. Nanomed. 2020, 15, 9823–9857. [Google Scholar] [CrossRef]
- Malathi, S.; Balakumaran, M.D.; Kalaichelvan, P.T.; Balasubramanian, S. Green Synthesis of Gold Nanoparticles for Controlled Delivery. Adv. Mater. Lett. 2013, 4, 933–940. [Google Scholar] [CrossRef]
- Majdi, H.; Salehi, R.; Pourhassan-Moghaddam, M.; Mahmoodi, S.; Poursalehi, Z.; Vasilescu, S. Antibody Conjugated Green Synthesized Chitosan-Gold Nanoparticles for Optical Biosensing. Colloids Interface Sci. Commun. 2019, 33, 100207. [Google Scholar] [CrossRef]
- Thanayutsiri, T.; Patrojanasophon, P.; Opanasopit, P.; Ngawhirunpat, T.; Plianwong, S.; Rojanarata, T. Rapid Synthesis of Chitosan-Capped Gold Nanoparticles for Analytical Application and Facile Recovery of Gold from Laboratory Waste. Carbohydr. Polym. 2020, 250, 116983. [Google Scholar] [CrossRef]
- Mi, X.; Choi, H.S.; Park, H.; Kim, Y.J. Structural Characterization and Anti-Inflammatory Properties of Green Synthesized Chitosan/Compound K-Gold Nanoparticles. Int. J. Biol. Macromol. 2022, 213, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.O.; Gunasekaran, S.; Ravishankar, C.N. Chitosan-Capped Gold Nanoparticles for Indicating Temperature Abuse in Frozen Stored Products. npj Sci. Food 2019, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, X. Synthesis of Chitosan-Stabilized Gold Nanoparticles in the Absence/Presence of Tripolyphosphate. Biomacromolecules 2004, 5, 2340–2346. [Google Scholar] [CrossRef] [PubMed]
- Koushki, E. Effect of Conjugation with Organic Molecules on the Surface Plasmon Resonance of Gold Nanoparticles and Application in Optical Biosensing. RSC Adv. 2021, 11, 23390–23399. [Google Scholar] [CrossRef]
- Oliveira, J.P.; Prado, A.R.; Keijok, W.J.; Antunes, P.W.P.; Yapuchura, E.R.; Guimarães, M.C.C. Impact of Conjugation Strategies for Targeting of Antibodies in Gold Nanoparticles for Ultrasensitive Detection of 17β-Estradiol. Sci. Rep. 2019, 9, 13859. [Google Scholar] [CrossRef]
- El-Sayed, I.H.; Huang, X.; El-Sayed, M.A. Surface Plasmon Resonance Scattering and Absorption of Anti-EGFR Antibody Conjugated Gold Nanoparticles in Cancer Diagnostics: Applications in Oral Cancer. Nano Lett. 2005, 5, 829–834. [Google Scholar] [CrossRef]
- Monti, S.; Jose, J.; Sahajan, A.; Kalarikkal, N.; Thomas, S. Structure and Dynamics of Gold Nanoparticles Decorated with Chitosan-Gentamicin Conjugates: ReaxFF Molecular Dynamics Simulations to Disclose Drug Delivery. Phys. Chem. Chem. Phys. 2019, 21, 13099–13108. [Google Scholar] [CrossRef]
- Barth, A. Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta Bioenergetics 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent Advancements in Optical Biosensors for Cancer Detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of Size and Concentration of Gold Nanoparticles from UV−Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef] [PubMed]
- Kasthuri, J.; Rajendiran, N. Functionalization of Silver and Gold Nanoparticles Using Amino Acid Conjugated Bile Salts with Tunable Longitudinal Plasmon Resonance. Colloids Surfaces B Biointerfaces 2009, 73, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Asal, H.A.; Shoueir, K.R.; El-Hagrasy, M.A.; Toson, E.A. Controlled Synthesis of In-Situ Gold Nanoparticles onto Chitosan Functionalized PLGA Nanoparticles for Oral Insulin Delivery. Int. J. Biol. Macromol. 2022, 209, 2188–2196. [Google Scholar] [CrossRef] [PubMed]
- Singpanna, K.; Pornpitchanarong, C.; Patrojanasophon, P.; Rojanarata, T.; Ngawhirunpat, T.; Li, S.K.; Opanasopit, P. Chitosan Capped-Gold Nanoparticles as Skin Penetration Enhancer for Small Molecules: A Study in Porcine Skin. Int. J. Pharm. 2023, 640, 123034. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, D.; Wang, Z. Synthesis and Cell-Surface Binding of Lectin-Gold Nanoparticle Conjugates. Anal. Methods 2011, 3, 1745. [Google Scholar] [CrossRef]
- Sitnikova, V.E.; Kotkova, M.A.; Nosenko, T.N.; Kotkova, T.N.; Martynova, D.M.; Uspenskaya, M.V. Breast Cancer Detection by ATR-FTIR Spectroscopy of Blood Serum and Multivariate Data-Analysis. Talanta 2020, 214, 120857. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1206. [Google Scholar] [CrossRef]
- Lewis, A.T.; Jones, K.; Lewis, K.E.; Jones, S.; Lewis, P.D. Detection of Lewis Antigen Structural Change by FTIR Spectroscopy. Carbohydr. Polym. 2013, 92, 1294–1301. [Google Scholar] [CrossRef]
- Nallala, J.; Jeynes, C.; Saunders, S.; Smart, N.; Lloyd, G.; Riley, L.; Salmon, D.; Stone, N. Characterization of Colorectal Mucus Using Infrared Spectroscopy: A Potential Target for Bowel Cancer Screening and Diagnosis. Lab. Investig. 2020, 100, 1102–1110. [Google Scholar] [CrossRef]
- Lewis, S.P.; Lewis, A.T.; Lewis, P.D. Prediction of Glycoprotein Secondary Structure Using ATR-FTIR. Vib. Spectrosc. 2013, 69, 21–29. [Google Scholar] [CrossRef]
- Rodriguez, P.E.A.; Maggio, B.; Cumar, F.A. Acid and Enzymatic Hydrolysis of the Internal Sialic Acid Residue in Native and Chemically Modified Ganglioside GM1. J. Lipid Res. 1996, 37, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Eid, S.M.; Kelani, K.M.; Badran, O.M.; Rezk, M.R.; Elghobashy, M.R. Surface Enhanced Infrared Absorption Spectroscopy (SEIRA) as a Green Analytical Chemistry Approach: Coating of Recycled Aluminum TLC Sheets with Citrate Capped Silver Nanoparticles for Chemometric Quantitative Analysis of Ternary Mixtures as a Green Alter. Anal. Chim. Acta 2020, 1117, 60–73. [Google Scholar] [CrossRef]
- Verger, F.; Pain, T.; Nazabal, V.; Boussard-Plédel, C.; Bureau, B.; Colas, F.; Rinnert, E.; Boukerma, K.; Compère, C.; Guilloux-Viry, M.; et al. Surface Enhanced Infrared Absorption (SEIRA) Spectroscopy Using Gold Nanoparticles on As2S3 Glass. Sens. Actuators B Chem. 2012, 175, 142–148. [Google Scholar] [CrossRef]
- Bibikova, O.; Haas, J.; López-Lorente, Á.I.; Popov, A.; Kinnunen, M.; Ryabchikov, Y.; Kabashin, A.; Meglinski, I.; Mizaikoff, B. Surface Enhanced Infrared Absorption Spectroscopy Based on Gold Nanostars and Spherical Nanoparticles. Anal. Chim. Acta 2017, 990, 141–149. [Google Scholar] [CrossRef]
- Kumar, N.; Thomas, S.; Tokas, R.B.; Kshirsagar, R.J. SEIRA Studies of Uracil Adsorbed on Wet-Chemically Prepared Gold Nanoparticles Film on Glass Substrate—Effect of Morphology of Film. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 129, 359–364. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Denizli, A. Plasmonic Nanosensors for Pharmaceutical and Biomedical Analysis. J. Pharm. Biomed. Anal. 2023, 236, 115671. [Google Scholar] [CrossRef]
- Song, W.; Ding, L.; Chen, Y.; Ju, H. Plasmonic Coupling of Dual Gold Nanoprobes for SERS Imaging of Sialic Acids on Living Cells. Chem. Commun. 2016, 52, 10640–10643. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate Classification for Vibrational Spectroscopy in Biological Samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef]
- Cadoret, M.; Husson, F. Construction and Evaluation of Confidence Ellipses Applied at Sensory Data. Food Qual. Prefer. 2013, 28, 106–115. [Google Scholar] [CrossRef]
- De Almeida, F.A.; Gomes, G.F.; Gaudêncio, J.H.D.; Gomes, J.H. de F.; Paiva, A.P. de A New Multivariate Approach Based on Weighted Factor Scores and Confidence Ellipses to Precision Evaluation of Textured Fiber Bobbins Measurement System. Precis. Eng. 2019, 60, 520–534. [Google Scholar] [CrossRef]
- Biswal, R.P.; Dandamudi, R.B.; Patnana, D.P.; Pandey, M.; Vutukuri, V.N.R.K. Metabolic Fingerprinting of Ganoderma Spp. Using UHPLC-ESI-QTOF-MS and Its Chemometric Analysis. Phytochemistry 2022, 199, 113169. [Google Scholar] [CrossRef] [PubMed]
- Dizaji, A.N.; Ozek, N.S.; Yilmaz, A.; Aysin, F.; Yilmaz, M. Gold Nanorod Arrays Enable Highly Sensitive Bacterial Detection via Surface-Enhanced Infrared Absorption (SEIRA) Spectroscopy. Colloids Surfaces B Biointerfaces 2021, 206, 111939. [Google Scholar] [CrossRef] [PubMed]
- de Graaff, D.R.; Felz, S.; Neu, T.R.; Pronk, M.; van Loosdrecht, M.C.M.; Lin, Y. Sialic Acids in the Extracellular Polymeric Substances of Seawater-Adapted Aerobic Granular Sludge. Water Res. 2019, 155, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.; Rani, S.; Kumar, V.; Nakhate, K.T.; Ajazuddin; Gupta, U. Sialic Acid Conjugated Chitosan Nanoparticles: Modulation to Target Tumour Cells and Therapeutic Opportunities. AAPS PharmSciTech 2022, 23, 10. [Google Scholar] [CrossRef] [PubMed]
- Kujdowicz, M.; Mech, B.; Chrabaszcz, K.; Chlosta, P.; Okon, K.; Malek, K. FTIR Spectroscopic Imaging Supports Urine Cytology for Classification of Low- and High-Grade Bladder Carcinoma. Cancers 2021, 13, 5734. [Google Scholar] [CrossRef]
- Medipally, D.K.R.; Cullen, D.; Untereiner, V.; Sockalingum, G.D.; Maguire, A.; Nguyen, T.N.Q.; Bryant, J.; Noone, E.; Bradshaw, S.; Finn, M.; et al. Vibrational Spectroscopy of Liquid Biopsies for Prostate Cancer Diagnosis. Ther. Adv. Med. Oncol. 2020, 12, 175883592091849. [Google Scholar] [CrossRef]
- Ghimire, H.; Garlapati, C.; Janssen, E.A.M.; Krishnamurti, U.; Qin, G.; Aneja, R.; Perera, A.G.U. Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis. Cancers 2020, 12, 1708. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zamudio Cañas, R.; Jaramillo Flores, M.E.; Vallejo Ruiz, V.; Delgado Macuil, R.J.; López Gayou, V. Detection of Sialic Acid to Differentiate Cervical Cancer Cell Lines Using a Sambucus nigra Lectin Biosensor. Biosensors 2024, 14, 34. https://doi.org/10.3390/bios14010034
Zamudio Cañas R, Jaramillo Flores ME, Vallejo Ruiz V, Delgado Macuil RJ, López Gayou V. Detection of Sialic Acid to Differentiate Cervical Cancer Cell Lines Using a Sambucus nigra Lectin Biosensor. Biosensors. 2024; 14(1):34. https://doi.org/10.3390/bios14010034
Chicago/Turabian StyleZamudio Cañas, Ricardo, María Eugenia Jaramillo Flores, Verónica Vallejo Ruiz, Raúl Jacobo Delgado Macuil, and Valentín López Gayou. 2024. "Detection of Sialic Acid to Differentiate Cervical Cancer Cell Lines Using a Sambucus nigra Lectin Biosensor" Biosensors 14, no. 1: 34. https://doi.org/10.3390/bios14010034