Carbon Nanotube-Supported Dummy Template Molecularly Imprinted Polymers for Selective Adsorption of Amide Herbicides in Aquatic Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals Reagents and Standard
2.2. Standard Solution Preparation
2.3. Preparation of Fish Samples and Spiked Samples
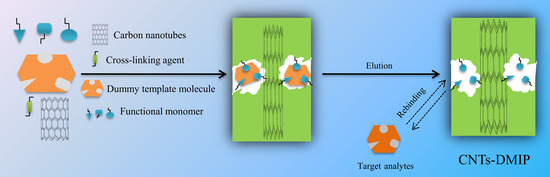
2.4. Preparation of CNTs@DMIPs
2.5. Sample Extraction and Clean-Up
2.6. Selective Adsorption of AmideH
2.7. LC-QqQMS
2.8. Data Analysis
3. Results
3.1. Characterization Results
3.2. Scatchard Analysis
3.3. Adsorption Kinetics
3.4. Selectivity
3.5. Extraction Conditions Optimisation
3.6. Matrix Effect
3.7. Method Validation
3.8. Applications to Fish Assays
3.9. Comparison with Other Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meseguer-Lloret, S.; Torres-Cartas, S.; Gómez-Benito, C.; Herrero-Martínez, J.M. Magnetic molecularly imprinted polymer for the simultaneous selective extraction of phenoxy acid herbicides from environmental water samples. Talanta 2022, 239, 123082. [Google Scholar] [CrossRef]
- Ahmadivand, S.; Farahmand, H.; Mirvaghefi, A.R.; Eagderi, S.; Shokrpoor, S.; Rahmati-Holasoo, H. Histopathological and haematological response of male rainbow trout (Oncorhynchus mykiss) subjected to butachlor. Vet. Med. Czech 2014, 59, 433–439. [Google Scholar] [CrossRef]
- Yu, J.X.; Xu, E.G.B.; Ren, Y.; Jin, S.Y.; Zhang, T.L.; Liu, J.S.; Li, Z.J. Mixture Toxicity of Bensulfuron-Methyl and Acetochlor to Red Swamp Crayfish (Procambarus clarkii): Behavioral, Morphological and Histological Effects. Int. J. Env. Res. Public Health 2017, 14, 1466. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Bai, X.; Zhang, T.; Yang, Z. Ultrasound-assisted extraction and solid-phase extraction for the simultaneous determination of five amide herbicides in fish samples by gas chromatography with electron capture detection. J. Sep. Sci. 2017, 40, 1142–1149. [Google Scholar] [CrossRef]
- Andrascikova, M.; Hrouzkova, S. Fast Preconcentration of Pesticide Residues in Oilseeds by Combination of QuEChERS with Dispersive Liquid-Liquid Microextraction Followed by Gas Chromatography-Mass Spectrometry. Food Anal. Methods 2016, 9, 2182–2193. [Google Scholar] [CrossRef]
- Orso, D.; Floriano, L.; Ribeiro, L.C.; Bandeira, N.M.G.; Prestes, O.D.; Zanella, R. Simultaneous Determination of Multiclass Pesticides and Antibiotics in Honey Samples Based on Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry. Food Anal. Methods 2016, 9, 1638–1653. [Google Scholar] [CrossRef]
- Li, Y.G.; Chen, Z.L.; Zhang, R.; Luo, P.; Zhou, Y.; Wen, S.; Ma, M.H. Simultaneous Determination of 42 Pesticides and Herbicides in Chicken Eggs by UHPLC-MS/MS and GC-MS Using a QuEChERS-Based Procedure. Chromatographia 2016, 79, 1165–1175. [Google Scholar] [CrossRef]
- Dong, X.F.; Liang, S.X.; Shi, Z.H.; Sun, H.W. Development of multi-residue analysis of herbicides in cereal grain by ultra-performance liquid chromatography-electrospray ionization-mass spectrometry. Food Chem. 2016, 192, 432–440. [Google Scholar] [CrossRef]
- Zou, N.; Han, Y.T.; Li, Y.J.; Qin, Y.H.; Gu, K.J.; Zhang, J.R.; Pan, C.P.; Li, X.S. Multiresidue Method for Determination of 183 Pesticide Residues in Leeks by Rapid Multiplug Filtration Cleanup and Gas Chromatography-Tandem Mass Spectrometry. J. Agr. Food Chem. 2016, 64, 6061–6070. [Google Scholar] [CrossRef]
- Qin, Y.H.; Zhang, J.R.; Zhang, Y.; Li, F.B.; Han, Y.T.; Zou, N.; Xu, H.W.; Qian, M.Y.; Pan, C.P. Automated multi-plug filtration cleanup for liquid chromatographic tandem mass spectrometric pesticide multi-residue analysis in representative crop commodities. J. Chromatogr. A 2016, 1462, 19–26. [Google Scholar] [CrossRef]
- Wu, L.J.; Li, Z.C. Continuous-flow microwave-assisted extraction coupled with online single drop microextraction prior to GC-MS for determination of amide herbicides in rice samples. J. Sep. Sci. 2021, 44, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.S.; Diao, C.P.; Chen, Q.F.; Wang, X. Sensitive determination of amide herbicides in environmental water samples by a combination of solid-phase extraction and dispersive liquid-liquid microextraction prior to GC-MS. J. Sep. Sci. 2009, 32, 1069–1074. [Google Scholar] [CrossRef]
- Gao, L.; Wang, P.; Chen, Z.X.; Hao, Q.R.; Bai, S.Y.; Du, N.N.; Li, C.H.; Huang, X.L.; Qin, D.L. Application of solid-phase extraction: High-resolution mass spectrometry analysis strategy in the characterization and quantification of amide herbicides in aquatic products. Electrophoresis 2022, 43, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Sun, Q.; Xu, B.; Yu, Y.; Wang, X. Molecularly imprinted solid phase extraction in a syringe filter for determination of triazine herbicides in Radix Paeoniae Alba by ultra-fast liquid chromatography. Talanta 2016, 148, 539–547. [Google Scholar] [CrossRef]
- Li, J.-X.; Sun, M.; Hu, X.-Y.; Wu, X.-Q.; Chang, Q.Y.; Fan, C.L. Rapid Determination of 11 Amide Herbicides in Tea by Headspace Solid-phase Microextraction Combined with Gas Chromatography-Triple Quadrupole Mass Spectrometry. J. Instrum. Anal. 2017, 11, 1339–1345. [Google Scholar] [CrossRef]
- Safari, M.; Yamini, Y. Application of magnetic nanomaterials in magnetic in-tube solid-phase microextraction. Talanta 2021, 221, 121648. [Google Scholar] [CrossRef] [PubMed]
- Kaczyński, P. Large-scale multi-class herbicides analysis in oilseeds by rapid one-step QuEChERS-based extraction and cleanup method using liquid chromatography–tandem mass spectrometry. Food Chem. 2017, 230, 411–422. [Google Scholar] [CrossRef]
- Liu, J.; Ji, C.H.; Liu, X.L.; Li, X.S.; Wu, H.Y.; Zeng, D.Q. Fe3O4 nanoparticles as matrix solid-phase dispersion extraction adsorbents for the analysis of thirty pesticides in vegetables by ultrahigh-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2021, 1165, 122532. [Google Scholar] [CrossRef]
- Zhang, L.; Han, F.; Hu, Y.; Zheng, P.; Sheng, X.; Sun, H.; Song, W.; Lv, Y. Selective trace analysis of chloroacetamide herbicides in food samples using dummy molecularly imprinted solid phase extraction based on chemometrics and quantum chemistry. Anal. Chim. Acta 2012, 729, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qiu, J.; Zhu, C.; Hua, Y.; Yu, J.; Jia, L.; Xu, J.; Li, J.; Li, Q. A Fluorescent Molecularly Imprinted Polymer-Coated Paper Sensor for On-Site and Rapid Detection of Glyphosate. Molecules 2023, 28, 2398. [Google Scholar] [CrossRef]
- Yang, J.-J.; Shen, Y.-Z.; Wang, Z.; Zhou, B.; Hu, X.-Y.; Xu, Q. β-Bi2O3 Nanosheets Functionalized with Bisphenol a Synthetic Receptors: A Novel Material for Sensitive Photoelectrochemical Platform Construction. Nanomaterials 2023, 13, 915. [Google Scholar] [CrossRef]
- Ye, L. Molecularly imprinted materials for bio/chemical analysis. Microchim. Acta 2023, 190, 31. [Google Scholar] [CrossRef]
- Farooq, S.; Chen, B.; Ahmad, S.; Muhammad, I.; Hussain, Q.; Wu, H. Room-Temperature, Ionic-Liquid-Enhanced, Beta-Cyclodextrin-Based, Molecularly Imprinted Polymers for the Selective Extraction of Abamectin. Nanomaterials 2022, 12, 1017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, J.; Sun, J.-S.; Li, Y.; Xue, S.-X.; Chen, X.-Q.; Li, X.-S.; Du, G.-X. Facile synthesis of multifunctional multi-walled carbon nanotube for pathogen Vibrio alginolyticus detection in fishery and environmental samples. Talanta 2014, 128, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Pundir, M.; Prasher, P.; Vasić, K.; Leitgeb, M.; Kumar, A.; Prakash, R.; Knez, Ž.; Pandey, J.K.; Kumar, S. Enzyme modified CNTs for biosensing application: Opportunities and challenges. Colloid Interface Sci. Commun. 2021, 44, 100506. [Google Scholar] [CrossRef]
- Chalmpes, N.; Kouloumpis, A.; Zygouri, P.; Karouta, N.; Spyrou, K.; Stathi, P.; Tsoufis, T.; Georgakilas, V.; Gournis, D.; Rudolf, P. Layer-by-Layer Assembly of Clay-Carbon Nanotube Hybrid Superstructures. ACS Omega 2019, 4, 18100–18107. [Google Scholar] [CrossRef]
- Patila, M.; Chalmpes, N.; Dounousi, E.; Stamatis, H.; Gournis, D. Chapter Twelve—Use of functionalized carbon nanotubes for the anobiocatalysts. Methods Enzymol. 2020, 630, 263–301. [Google Scholar] [CrossRef]
- Lim, C.C.; Shuit, S.H.; Ng, Q.H.; Ab Rahim, S.K.E.; Hoo, P.Y.; Yeoh, W.M.; Goh, S.W. Sulfonated magnetic multi-walled carbon nanotubes with enhanced bonding stability, high adsorption performance, and reusability for water remediation. Environ. Sci. Pollut. Res. 2023, 30, 40242–40259. [Google Scholar] [CrossRef]
- Solorio-Rodriguez, S.A.; Williams, A.; Poulsen, S.S.; Knudsen, K.B.; Jensen, K.A.; Clausen, P.A.; Danielsen, P.H.; Wallin, H.; Vogel, U.; Halappanavar, S. Single-Walled vs. Multi-Walled Carbon Nanotubes: Influence of Physico-Chemical Properties on Toxicogenomics Responses in Mouse Lungs. Nanomaterials 2023, 13, 1059. [Google Scholar] [CrossRef]
- Gao, L.; We, Z.; Qin, D.; Chen, Z.; Wang, P. Challenges and Prospects of Detection Technology for Organic Pollutant Residues in Fishery Products. Chin. J. Fish. 2021, 34, 82–86. [Google Scholar]
- Gao, L.; Qin, D.; Chen, Z.; Wu, S.; Tang, S.; Wang, P. Determination of sulfonamide antibiotics in fish and shrimp samples based on magnetic carbon nanotube dummy molecularly imprinted polymer extraction followed by UPLC-MS/MS. Electrophoresis 2021, 42, 725–734. [Google Scholar] [CrossRef]
- Liu, H.C.; Hong, Y.S.; Chen, L.G. Molecularly imprinted polymers coated on carbon nanotubes for matrix solid phase dispersion extraction of camptothecin from Camptotheca acuminate. Anal. Methods 2015, 7, 8100–8108. [Google Scholar] [CrossRef]
- Ni, R.; Wang, Y.; Wei, X.; Chen, J.; Meng, J.; Xu, F.; Liu, Z.; Zhou, Y. Magnetic carbon nanotube modified with polymeric deep eutectic solvent for the solid phase extraction of bovine serum albumin. Talanta 2020, 206, 120215. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, Y.-S.; Ren, X.-H.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. Bimetallic molecularly imprinted nanozyme: Dual-mode detection platform. Biosens. Bioelectron. 2022, 196, 113718. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, X.; Wu, M.; Ma, Y.; Xu, X.; Chen, L.; Niu, N. Facile off-on fluorescence biosensing of human papillomavirus using DNA probe coupled with sunflower seed shells carbon dots. Microchem. J. 2022, 181, 107742. [Google Scholar] [CrossRef]
- Deeyai, P.; Suphantharika, M.; Wongsagonsup, R.; Dangtip, S. Characterization of Modified Tapioca Starch in Atmospheric Argon Plasma under Diverse Humidity by FTIR Spectroscopy. Chin. Phys. Lett. 2013, 30, 018103. [Google Scholar] [CrossRef]
- Prabakaran, K.; Jandas, P.J.; Luo, J.T.; Fu, C.; Wei, Q.P. Molecularly imprinted poly(methacrylic acid) based QCM biosensor for selective determination of L-tryptophan. Colloid Interface Sci. Commun. 2021, 611, 125859. [Google Scholar] [CrossRef]
- Sun, Y.C.; Bai, L.; Han, C.H.; Lv, X.T.; Sun, X.Y.; Wang, T.T. Hybrid amino-functionalized TiO2/sodium lignosulfonate surface molecularly imprinted polymer for effective scavenging of methylene blue from wastewater. J. Clean. Prod. 2022, 337, 130457. [Google Scholar] [CrossRef]
- Kuang, Y.; Chen, W.; Chen, Z.H.; Xia, Y.L.; Rao, Q.Q.; Yang, S.X. Highly-efficient selective recognition and rapid enrichment of chrysin by magnetic surface molecularly imprinted polymer. Food Chem. 2023, 405, 134993. [Google Scholar] [CrossRef]





| Recognized Molecule | CNTs@DMIPs | CNTs@DNIPs | β | ||
|---|---|---|---|---|---|
| δ | α | δ | α | ||
| alachlor | 82.20 | 10.72 | 21.57 | 5.14 | 2.09 |
| acetochlor | 68.33 | 8.91 | 29.53 | 7.03 | 1.27 |
| pretilachlor | 72.80 | 9.49 | 20.80 | 4.95 | 1.92 |
| butachlor | 73.53 | 9.59 | 21.17 | 5.04 | 1.90 |
| metolachlor | 81.70 | 10.65 | 17.23 | 4.10 | 2.60 |
| diethatylethyl | 79.90 | 10.42 | 15.73 | 3.75 | 2.78 |
| dimethachlor | 91.80 | 11.97 | 27.77 | 6.61 | 1.81 |
| atrazine | 7.67 | - | 4.20 | - | - |
| Amide Herbicides | Linearity μgkg−1 | LOD μgkg−1 | LOQ μgkg−1 | Spiked Level 0.05 μgkg−1 | Spiked Level 1.0 μgkg−1 | Spiked Level 2.0 μgkg−1 | |||
|---|---|---|---|---|---|---|---|---|---|
| Recovery (%) | RSD (%) | Recovery (%) | RSD (%) | Recovery (%) | RSD (%) | ||||
| alachlor | 0.05–2.5 | 0.025 | 0.05 | 95.83 | 4.16 | 94.68 | 1.58 | 94.74 | 4.52 |
| acetochlor | 0.025–2.5 | 0.0125 | 0.025 | 98.90 | 3.35 | 93.02 | 3.31 | 104.02 | 5.60 |
| pretilachlor | 0.025–2.5 | 0.0125 | 0.025 | 100.70 | 2.55 | 96.47 | 3.77 | 102.51 | 4.79 |
| butachlor | 0.05–2.5 | 0.025 | 0.05 | 90.74 | 6.28 | 87.34 | 3.97 | 86.90 | 4.74 |
| metolachlor | 0.025–2.5 | 0.0125 | 0.025 | 94.65 | 4.36 | 86.96 | 3.85 | 98.93 | 1.31 |
| diethatyl ethyl | 0.05–2.5 | 0.025 | 0.05 | 98.55 | 6.55 | 101.47 | 3.58 | 100.02 | 1.58 |
| dimethachlor | 0.025–2.5 | 0.0125 | 0.025 | 94.92 | 6.30 | 91.12 | 4.33 | 93.23 | 6.14 |
| Extraction | Clean-Up | Determination | Sample | Analytical | LOD (μgkg−1) | Reference |
|---|---|---|---|---|---|---|
| Dispersive liquid-liquid microextraction | QuEChERS | GC-MS | Oilseeds | Alachlor, acetochlor | 0.07–0.7 | [5] |
| Dispersive SPE | SPE | LC-MS/MS | Honey | Metolachlor | 0.1 | [6] |
| Shaking | QuEChERS | LC-MS/MS | Chicken eggs | Butachlor, acetochlor | 0.2 | [7] |
| Shaking | SPE | LC-MS | Cereal grain | Metazachlor | 0.08 | [8] |
| Dispersive SPE | Multiplug filtration clean-up. | GC-MS/MS | Leeks | Alachlor, butachlor, acetochlor | 1.7–2.6 | [9] |
| Reversed-dispersive SPE | Multiplug filtration clean-up. | LC-MS/MS | Peanut | Acetochlor | 2.0 | [10] |
| MSPD | CNTs@DMIPs | LC-MS/MS | Fish | Alachlor, acetochlor, pretilachlor, butachlor, metolachlor, diethatyl ethyl, dimethachlor | 0.0125– 0.025 | Proposed method |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, S.; Li, C.; Huang, L.; Chen, Z.; Wang, P.; Qin, D.; Gao, L. Carbon Nanotube-Supported Dummy Template Molecularly Imprinted Polymers for Selective Adsorption of Amide Herbicides in Aquatic Products. Nanomaterials 2023, 13, 1521. https://doi.org/10.3390/nano13091521
Zeng S, Li C, Huang L, Chen Z, Wang P, Qin D, Gao L. Carbon Nanotube-Supported Dummy Template Molecularly Imprinted Polymers for Selective Adsorption of Amide Herbicides in Aquatic Products. Nanomaterials. 2023; 13(9):1521. https://doi.org/10.3390/nano13091521
Chicago/Turabian StyleZeng, Sili, Chenhui Li, Li Huang, Zhongxiang Chen, Peng Wang, Dongli Qin, and Lei Gao. 2023. "Carbon Nanotube-Supported Dummy Template Molecularly Imprinted Polymers for Selective Adsorption of Amide Herbicides in Aquatic Products" Nanomaterials 13, no. 9: 1521. https://doi.org/10.3390/nano13091521