Graphene Decorated Zinc Oxide and Curcumin to Disinfect the Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Reduced Gr-ZnO Nanocomposite
2.2. Characterization of As-Synthesized GrZnO-NCs
2.2.1. UV-Visible Spectroscopy
2.2.2. Dynamic Light Scattering
2.2.3. Electron Microscopy
2.2.4. X-ray Diffraction Analysis
2.3. Minimum Inhibitory Concentration (M.I.C.) and Antibacterial Efficacy Determined through Agar Diffusion Assay
2.4. The Synergistic Anti-Microbial Effect of C.C.M. Supplemented with GrZnO-NCs Determined by the Agar Diffusion Method
2.5. M.R.S.A. Viability Assay Employing SYTO9 and Propidium Iodide (P.I.) Dyes
2.6. Anti-Biofilm Activity
2.7. Bacterial Susceptibility Against As-Synthesized GrZnO-NCs
2.8. The Interaction between Bacteria -N.C.s Showed by Electron Microscopy
2.9. Determination of Cell Lyses by Protein Leakage
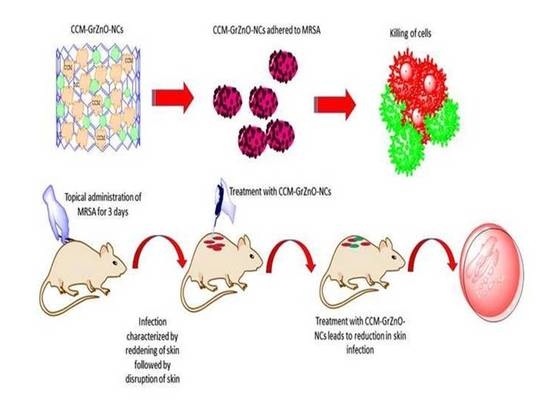
2.10. Potential of GrZnO-NCs in the Suppression of Experimental M.R.S.A. Skin Infection
2.11. Statistical Analysis
3. Results and Discussion
3.1. Characterization of As-Synthesized GrZnO-NCs
3.2. The Synergistic Antibacterial Potential of As-Synthesized Reduced GrZnO-NCs in Combination with CCM
3.3. The Anti-Biofilm Potential of As-Synthesized GrZnO-NCs
3.4. Influence of CCM-GrZnO-NCs Formulation on Bacteria as Depicted by E.M. Examination
3.5. Histopathological Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Emerging infectious diseases of wildlife—Threats to biodiversity and human health. Science 2000, 287, 443. [Google Scholar] [CrossRef] [PubMed]
- Oaks, S.C., Jr.; Shope, R.E.; Lederberg, J. Emerging Infections: Microbial Threats to Health in the United States; National Academies Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Schweizer, H.P. Bacterial resistance to antibiotics: Active efflux and reduced uptake. Adv. Drug Deliv. Rev. 2005, 57, 1486–1513. [Google Scholar] [CrossRef]
- Huh, A.J.; Kwon, Y.J. Nanoantibiotics: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Qin, L.; Zeng, G.; Lai, C.; Huang, D.; Xu, P.; Zhang, C.; Cheng, M.; Liu, X.; Liu, S.; Li, B.; et al. “Gold rush” in modern science: Fabrication strategies and typical advanced applications of gold nanoparticles in sensing. Coord. Chem. Rev. 2018, 359, 1–31. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghupathi, K.R.; Koodali, R.T.; Manna, A.C. Size-dependent bacterial growth inhibition and mechanism of antibacterial activity of zinc oxide nanoparticles. Langmuir 2011, 27, 4020–4028. [Google Scholar] [CrossRef]
- Oves, M.; Aslam, M.; Rauf, M.A.; Qayyum, S.; Qari, H.A.; Khan, M.S.; Alam, M.Z.; Tabrez, S.; Pugazhendhi, A.; Ismail, I.M. Anti-microbial and anticancer activities of silver nanoparticles synthesized from the root hair extract of Phoenix dactylifera. Mater. Sci. Eng. C 2018, 89, 429–443. [Google Scholar] [CrossRef]
- Pati, R.; Mehta, R.K.; Mohanty, S.; Padhi, A.; Sengupta, M.; Vaseeharan, B.; Goswami, C.; Sonawane, A. Topical application of zinc oxide nanoparticles reduces bacterial skin infection in mice and exhibits antibacterial activity by inducing oxidative stress response and cell membrane disintegration in macrophages. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1195–1208. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Fromm, K.M.; Ashkarran, A.A.; de Aberasturi, D.J.; de Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Bai, R.G.; Muthoosamy, K.; Shipton, F.N.; Manickam, S. Acoustic cavitation induced generation of stabilizer-free, extremely stable reduced graphene oxide nanodispersion for efficient delivery of paclitaxel in cancer cells. Ultrason. Sonochem. 2017, 36, 129–138. [Google Scholar]
- Hummers, W.S., Jr.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Zhang, Z.; Ortiz, O.; Goyal, R.; Kohn, J. Chapter 23—Biodegradable Polymers. In Principles of Tissue Engineering, 4th ed.; Lanza, R., Langer, R., Vacanti, R., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 441–473. [Google Scholar] [CrossRef]
- Miriyala, S.; Panchatcharam, M.; Rengarajulu, P. Cardioprotective Effects of Curcumin. In The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2007; pp. 359–377. [Google Scholar]
- Akram, M.; Shahab-Uddin, A.A.; Usmanghani, K.; Hannan, A.; Mohiuddin, E.; Asif, M. Curcuma longa and curcumin: A review article. Rom J. Biol. Plant Biol. 2010, 55, 65–70. [Google Scholar]
- Mun, S.H.; Kim, S.B.; Kong, R.; Choi, J.G.; Kim, Y.C.; Shin, D.W.; Kang, O.H.; Kwon, D.Y. Curcumin reverse methicillin resistance in Staphylococcus aureus. Molecules 2014, 19, 18283–18295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauf, M.A.; Zubair, S.; Ateeq, H.; Dabeer, K.; Pachauri, S.; Ajmal, M.; Owais, M. Synergistic effect of Diallyl sulphide with Zinc oxide Nanorods: A novel and effective approach for treatment of acute dermatitis in model animals. Front. Microbiol. 2018, 9, 586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packiavathy, I.A.S.V.; Sasikumar, P.; Pandian, S.K.; Ravi, A.V. Prevention of quorum-sensing-mediated biofilm development and virulence factors production in Vibrio spp. by Curcumin. Appl. Microbiol. Biotechnol. 2013, 97, 10177–10187. [Google Scholar] [CrossRef] [PubMed]
- Teow, S.Y.; Liew, K.; Ali, S.A.; Khoo, A.S.B.; Peh, S.C. Antibacterial action of Curcumin against Staphylococcus aureus: A brief review. J. Trop. Med. 2016, 2016, 2853045. [Google Scholar] [CrossRef] [Green Version]
- Zorofchian Moghadamtousi, S.; Abdul Kadir, H.; Hassandarvish, P.; Tajik, H.; Abubakar, S.; Zandi, K. A review on antibacterial, antiviral, and antifungal activity of Curcumin. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Chen, J.; Yao, B.; Li, C.; Shi, G. An improved Hummers method for eco-friendly synthesis of graphene oxide. Carbon 2013, 64, 225–229. [Google Scholar] [CrossRef]
- Zhang, Q.; Tian, C.; Wu, A.; Tan, T.; Sun, L.; Wang, L.; Fu, H. A facile one-pot route for the controllable growth of small sized and well-dispersed ZnO particles on GO-derived graphene. J. Mater. Chem. 2012, 22, 11778–11784. [Google Scholar] [CrossRef]
- Stankovich, S.; Dikin, D.A.; Piner, R.D.; Kohlhaas, K.A.; Kleinhammes, A.; Jia, Y.; Wu, Y.; Nguyen, S.T.; Ruoff, R.S. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon 2007, 45, 1558–1565. [Google Scholar] [CrossRef]
- Chua, C.K.; Pumera, M. Chemical reduction of graphene oxide: A synthetic chemistry viewpoint. Chem. Soc. Rev. 2014, 43, 291–312. [Google Scholar] [CrossRef] [PubMed]
- Muthoosamy, K.; Bai, R.G.; Abubakar, I.B.; Sudheer, S.M.; Lim, H.N.; Loh, H.S.; Huang, N.M.; Chia, C.H.; Manickam, S. Exceedingly biocompatible and thin-layered reduced graphene oxide nanosheets using an eco-friendly mushroom extract strategy. Int. J. Nanomed. 2015, 10, 1505. [Google Scholar]
- Muthoosamy, K.; GBai, R.; Manickam, S. Graphene and graphene oxide as a docking station for modern drug delivery system. Curr. Drug Deliv. 2014, 11, 701–718. [Google Scholar] [CrossRef]
- Mun, S.H.; Joung, D.K.; Kim, Y.S.; Kang, O.H.; Kim, S.B.; Seo, Y.S.; Kim, Y.C.; Lee, D.S.; Shin, D.W.; Kweon, K.T.; et al. Synergistic antibacterial effect of Curcumin against methicillin-resistant Staphylococcus aureus. Phytomedicine 2013, 20, 714–718. [Google Scholar] [CrossRef]
- Guess, W.L.; Rosenbluth, S.A.; Schmidt, B.; Autian, J. Agar diffusion method for toxicity screening of plastics on cultured cell monolayers. J. Pharm. Sci. 1965, 54, 1545–1547. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Khan, S.; Meena, R.; Singh, B.R.; Khan, A.U. A graphene/zinc oxide nanocomposite film protects dental implant surfaces against cariogenic Streptococcus mutans. Biofouling 2014, 30, 1281–1294. [Google Scholar] [CrossRef]
- Williams, S.C.; Hong, Y.; Danavall, D.C.A.; Howard-Jones, M.H.; Gibson, D.; Frischer, M.E.; Verity, P.G. Distinguishing between living and nonliving bacteria: Evaluation of the vital stain propidium iodide and its combined use with molecular probes in aquatic samples. J. Microbiol. Methods 1998, 32, 225–236. [Google Scholar] [CrossRef]
- Xu, Z.; Liang, Y.; Lin, S.; Chen, D.; Li, B.; Li, L.; Deng, Y. Crystal Violet and X.T.T. Assays on Staphylococcus aureus biofilm quantification. Curr. Microbiol. 2016, 73, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.A.; Owais, M.; Rajpoot, R.; Ahmad, F.; Khan, N.; Zubair, S. Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals. RSC Adv. 2017, 7, 36361–36373. [Google Scholar] [CrossRef] [Green Version]
- Stoscheck, C.M. Quantitation of protein. Methods Enzymol. 1990, 182, 50–68. [Google Scholar] [PubMed]
- Zhao, R.; Kong, W.; Sun, M.; Yang, Y.; Liu, W.; Lv, M.; Song, S.; Wang, L.; Song, H.; Hao, R. Highly Stable Graphene-Based Nanocomposite (G.O.–P.E.I.–Ag) with Broad-Spectrum, Long-Term Anti-microbial Activity and Antibiofilm Effects. ACS Appl. Mater. Interfaces 2018, 10, 17617–17629. [Google Scholar] [CrossRef] [PubMed]
- Marcano, D.C.; Kosynkin, D.V.; Berlin, J.M.; Sinitskii, A.; Sun, Z.; Slesarev, A.; Alemany, L.B.; Lu, W.; Tour, J.M. Improved synthesis of graphene oxide. ACS Nano 2010, 4, 4806–4814. [Google Scholar] [CrossRef]
- Shoeb, M.; Singh, B.R.; Mobin, M.; Afreen, G.; Khan, W.; Naqvi, A.H. Kinetic study on mutagenic chemical degradation through three pot synthesiszed graphene@ ZnO nanocomposite. PLoS ONE 2015, 10, e0135055. [Google Scholar] [CrossRef] [Green Version]
- Bai, R.G.; Muthoosamy, K.; Shipton, F.N.; Pandikumar, A.; Rameshkumar, P.; Huang, N.M.; Manickam, S. The biogenic synthesis of a reduced graphene oxide–silver (R.G.O.–Ag) nanocomposite and its dual applications as an antibacterial agent and cancer biomarker sensor. RSC Adv. 2016, 6, 36576–36587. [Google Scholar] [CrossRef]
- Chen, Y.L.; Hu, Z.A.; Chang, Y.Q.; Wang, H.W.; Zhang, Z.Y.; Yang, Y.Y.; Wu, H.Y. Zinc oxide/reduced graphene oxide composites and electrochemical capacitance enhanced by homogeneous incorporation of reduced graphene oxide sheets in zinc oxide matrix. J. Phys. Chem. C 2011, 115, 2563–2571. [Google Scholar] [CrossRef]
- Dikin, D.A.; Stankovich, S.; Zimney, E.J.; Piner, R.D.; Dommett, G.H.; Evmenenko, G.; Nguyen, S.T.; Ruoff, R.S. Preparation and characterization of graphene oxide paper. Nature 2007, 448, 457. [Google Scholar] [CrossRef]
- Raoufi, D. Synthesis and Synthesis and microstructural properties of ZnO nanoparticles prepared by precipitation method. Renew. Energy 2013, 50, 932–937. [Google Scholar] [CrossRef]
- Parveen, N.; Mahato, N.; Ansari, M.O.; Cho, M.H. Enhanced electrochemical behavior and hydrophobicity of crystalline polyaniline@graphene nanocomposite synthesized at elevated temperature. Compos. Part B Eng. 2016, 87, 281–290. [Google Scholar] [CrossRef]
- Dai, K.; Lu, L.; Liang, C.; Dai, J.; Zhu, G.; Liu, Z.; Liu, Q.; Zhang, Y. Graphene oxide modified ZnO nanorods hybrid with high reusable photocatalytic activity under UV-LED irradiation. Mater. Chem. Phys. 2014, 143, 1410–1416. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (M.I.C.) of anti-microbial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Cao, A.; Jiang, Y.; Zhang, X.; Liu, J.H.; Liu, Y.; Wang, H. Superior antibacterial activity of zinc oxide/graphene oxide composites originating from high zinc concentration localized around bacteria. ACS Appl. Mater. Interfaces 2014, 6, 2791–2798. [Google Scholar] [CrossRef] [PubMed]
- Varaprasad, K.; Mohan, Y.M.; Vimala, K.; Mohana Raju, K. Synthesis and characterization of hydrogel silver nanoparticle curcumin composites for wound dressing and antibacterial application. J. Appl. Polym. Sci. 2011, 121, 784–796. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Z.; Wu, H.; Lv, F. Study on the antibiotic activity of microcapsule curcumin against foodborne pathogens. Int. J. Food Microbiol. 2009, 136, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef]
- Bhise, K.; Sau, S.; Kebriaei, R.; Rice, S.A.; Stamper, K.C.; Alsaab, H.O.; Rybak, M.J.; Iyer, A.K. Combination of vancomycin and cefazolin lipid nanoparticles for overcoming antibiotic resistance of MRSA. Materials 2018, 11, 1245. [Google Scholar] [CrossRef] [Green Version]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95. [Google Scholar] [CrossRef]
- Alam, M.; Dwivedi, V.; Khan, A.A.; Mohammad, O. Efficacy of niosomal formulation of diallyl sulfide against experimental candidiasis in Swiss albino mice. Nanomedicine 2009, 4, 713–724. [Google Scholar] [CrossRef]
- Ramos, F.A.; Takaishi, Y.; Shirotori, M.; Kawaguchi, Y.; Tsuchiya, K.; Shibata, H.; Higuti, T.; Tadokoro, T.; Takeuchi, M. Antibacterial and antioxidant activities of quercetin oxidation products from yellow onion (Allium cepa) skin. J. Agric. Food Chem. 2006, 54, 3551–3557. [Google Scholar] [CrossRef] [PubMed]









| Strains | GrZnO-NCs | CCM | CCM-GrZnO-NCs | Van (30 µg/disk) |
|---|---|---|---|---|
| ATCC 43300 | 14.6 ± 2 | 10.33 ± 2.082 | 21.33 ± 2.517 | 14.333 ± 2 |
| ATCC, BAA-1708 | 13.3 ± 1.5 | 9 ± 2 | 18.33 ± 0.677 | 9.67 ± 2.5 |
| Pathogen | Groups | Log10 CFU/mL |
|---|---|---|
| MRSA 43300 | Positive Control | 6.17 |
| GrZnO-NCs | 4.393 | |
| CCM | 4.773 | |
| CCM-GrZnO-NCs | 3.3 | |
| Vancomycin | 4.17 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oves, M.; Rauf, M.A.; Ansari, M.O.; Aslam Parwaz Khan, A.; A Qari, H.; Alajmi, M.F.; Sau, S.; Iyer, A.K. Graphene Decorated Zinc Oxide and Curcumin to Disinfect the Methicillin-Resistant Staphylococcus aureus. Nanomaterials 2020, 10, 1004. https://doi.org/10.3390/nano10051004
Oves M, Rauf MA, Ansari MO, Aslam Parwaz Khan A, A Qari H, Alajmi MF, Sau S, Iyer AK. Graphene Decorated Zinc Oxide and Curcumin to Disinfect the Methicillin-Resistant Staphylococcus aureus. Nanomaterials. 2020; 10(5):1004. https://doi.org/10.3390/nano10051004
Chicago/Turabian StyleOves, Mohammad, Mohd. Ahmar Rauf, Mohammad Omaish Ansari, Aftab Aslam Parwaz Khan, Huda A Qari, Mohamed F. Alajmi, Samaresh Sau, and Arun K Iyer. 2020. "Graphene Decorated Zinc Oxide and Curcumin to Disinfect the Methicillin-Resistant Staphylococcus aureus" Nanomaterials 10, no. 5: 1004. https://doi.org/10.3390/nano10051004