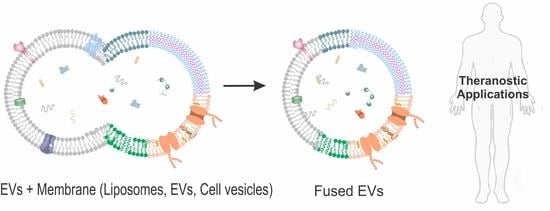
Tuning the Extracellular Vesicles Membrane through Fusion for Biomedical Applications
Abstract
:1. Introduction
2. Strategies of Membrane Fusion
2.1. pH-Mediated Fusion
2.2. Freeze–Thaw-Cycle-Mediated Fusion
2.3. Extrusion-Mediated Fusion
2.4. Polyethylene Glycol-Mediated Fusion
2.5. Natural Incubation
3. EVs and Fusion Membranes
3.1. EVs Fusion with Liposomes
3.2. EVs Fusion with EVs
3.3. EVs Fusion with Cell-Derived Membranes

4. Biomedical Applications of Fused EVs
4.1. EVs Fusion for Diagnostic Applications
4.2. EVs Fusion for Therapeutic Applications
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, S.; Zocco, D.; Samuels, M.L.; Chou, M.F.; Chammas, R.; Skog, J.; Zarovni, N.; Momen-Heravi, F.; Kuo, W.P. Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Chen, X.; Rechavi, O. Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 2022, 23, 185–203. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Budnik, V.; Ruiz-Cañada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology functionand biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Cheng, L.; Hill, A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022, 21, 379–399. [Google Scholar] [CrossRef]
- Bordanaba-Florit, G.; Royo, F.; Kruglik, S.G.; Falcón-Pérez, J.M. Using single-vesicle technologies to unravel the heterogeneity of extracellular vesicles. Nat. Protoc. 2021, 16, 3163–3185. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Lötvall, J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat. Protoc. 2021, 16, 1548–1580. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2021, 8, 2003505. [Google Scholar] [CrossRef] [PubMed]
- Roefs, M.T.; Sluijter, J.P.G.; Vader, P. Extracellular Vesicle-Associated Proteins in Tissue Repair. Trends Cell Biol. 2020, 30, 990–1013. [Google Scholar] [CrossRef] [PubMed]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Huang, Z.; Yang, Z.; Lin, Q.; Yang, B.; Fang, X.; Liu, B.; Chen, H.; Kong, J. Recent Progress in Detection and Profiling of Cancer Cell-Derived Exosomes. Nano Micro Small 2021, 17, 2007971. [Google Scholar] [CrossRef]
- Weber, T.; Zemelman, B.V.; McNew, J.A.; Westermann, B.; Gmachl, M.; Parlati, F.; Söllner, T.H.; Rothman, J.E. SNAREpins: Minimal Machinery for Membrane Fusion. Cell 1998, 92, 759–772. [Google Scholar] [CrossRef] [Green Version]
- Jahn, R.; Scheller, R.H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Stein, A.; Radhakrishnan, A.; Riedel, D.; Fasshauer, D.; Jahn, R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat. Struct. Mol. Biol. 2007, 14, 904–911. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.-H.M.; van Lengerich, B.; Boxer, S.G. Effects of Linker Sequences on Vesicle Fusion Mediated by Lipid-Anchored DNA Oligonucleotides. PNAS 2009, 106, 979–984. Available online: https://www.pnas.org/doi/abs/10.1073/pnas.0812356106 (accessed on 27 January 2009.). [CrossRef] [Green Version]
- Rørvig-Lund, A.; Bahadori, A.; Semsey, S.; Bendix, P.M.; Oddershede, L.B. Vesicle Fusion Triggered by Optically Heated Gold Nanoparticles. Nano Lett. 2015, 15, 4183–4188. [Google Scholar] [CrossRef]
- Connor, J.; Yatvin, M.B.; Huang, L. pH-Sensitive Liposomes: Acid-Induced Liposome Fusion. PNAS 1984, 81, 1715–1718. Available online: https://www.pnas.org/doi/abs/10.1073/pnas.81.6.1715 (accessed on 1 March 1984.). [CrossRef] [PubMed] [Green Version]
- Martens, S.; McMahon, H.T. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008, 9, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witkowska, A.; Heinz, L.P.; Grubmüller, H.; Jahn, R. Tight docking of membranes before fusion represents a metastable state with unique properties. Nat. Commun. 2021, 12, 3606. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- de Abreu, R.C.; Fernandes, H.; da Costa Martins, P.A.; Sahoo, S.; Emanueli, C.; Ferreira, L. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat. Rev. Cardiol. 2020, 17, 685–697. [Google Scholar] [CrossRef]
- Cavalcanti, R.R.M.; Lira, R.B.; Riske, K.A. Membrane Fusion Biophysical Analysis of Fusogenic Liposomes. Langmuir 2022, 38, 10430–10441. [Google Scholar] [CrossRef]
- Gao, F.; Yang, D.; Xu, F.; Ma, X.; Wang, P. Promoting Cell Fusion by Polyvalent DNA Ligands. Nano Lett. 2022, 22, 3018–3025. [Google Scholar] [CrossRef]
- Rahman, M.M.; Abosheasha, M.A.; Ito, Y.; Ueda, M. DNA-induced fusion between lipid domains of peptide–lipid hybrid vesicles. Chem. Commun. 2022, 58, 11799–11802. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Duan, R.; Zhou, Z.; Vázquez-González, M.; Xia, F.; Willner, I. Near-infrared light-activated membrane fusion for cancer cell therapeutic applications. Chem. Sci. 2020, 11, 5592–5600. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, Y.G.; Risselada, H.J.; Müller, M. Thermodynamically Reversible Paths of the First Fusion Intermediate Reveal An Important Role for Membrane Anchors of Fusion Proteins. PNAS 2019, 116, 2571–2576. Available online: https://www.pnas.org/doi/abs/10.1073/pnas.1818200116 (accessed on 30 January 2019). [CrossRef] [PubMed] [Green Version]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deli. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechno. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Millard, M.; Yakavets, I.; Piffoux, M.; Brun, A.; Gazeau, F.; Guigner, J.M.; Jasniewski, J.; Lassalle, H.P.; Wilhelm, C.; Bezdetnaya, L. mTHPC-loaded extracellular vesicles outperform liposomal and free mTHPC formulations by an increased stability, drug delivery efficiency and cytotoxic effect in tridimensional model of tumors. Drug Deliv. 2018, 25, 1790–1801. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Chen, Y.; Shi, J. Exosome Biochemistry and Advanced Nanotechnology for Next-Generation Theranostic Platforms. Adv. Mater 2019, 31, 1802896. [Google Scholar] [CrossRef]
- Liu, A.; Yang, G.; Liu, Y.; Liu, T. Research progress in membrane fusion-based hybrid exosomes for drug delivery systems. Front. Bioen. Biotechnol. 2022, 10, 939441. [Google Scholar] [CrossRef]
- Mondal, J.; Pillarisetti, S.; Junnuthula, V.; Saha, M.; Hwang, S.R.; Park, I.-k.; Lee, Y.-k. Hybrid exosomes, exosome-like nanovesicles and engineered exosomes for therapeutic applications. J. Control. Release 2023, 353, 1127–1149. [Google Scholar] [CrossRef]
- 51Akimov, S.A.; Molotkovsky, R.J.; Galimzyanov, T.R.; Radaev, A.V.; Shilova, L.A.; Kuzmin, P.I.; Batishchev, O.V.; Voronina, G.F.; Chizmadzhev, Y.A. Model of membrane fusion: Continuous transition to fusion pore with regard of hydrophobic and hydration interactions. Biochem. (Mosc.) Suppl. A Membr. Cell Biol. 2014, 8, 153–161. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Zimmerberg, J.; Kozlov, M.M. Membranes of the world unite! J. Cell Biol. 2006, 175, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Raphael, R.M. Solution pH Alters Mechanical and Electrical Properties of Phosphatidylcholine Membranes: Relation between Interfacial Electrostatics, Intramembrane Potential, and Bending Elasticity. Biophy. J. 2007, 92, 2451–2462. [Google Scholar] [CrossRef] [Green Version]
- Mitkova, D.; Vitkova, V. The aqueous surroundings alter the bending rigidity of lipid membranes. Russ. J. Electrochem. 2016, 52, 1172–1178. [Google Scholar] [CrossRef]
- Akimov, S.A.; Polynkin, M.A.; Jiménez-Munguía, I.; Pavlov, K.V.; Batishchev, O.V. Phosphatidylcholine Membrane Fusion Is pH-Dependent. Int. J. Mol. Sci. 2018, 19, 1358. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hong, Y.; Nam, G.-H.; Chung, J.H.; Koh, E.; Kim, I.-S. Virus-Mimetic Fusogenic Exosomes for Direct Delivery of Integral Membrane Proteins to Target Cell Membranes. Adv. Mater. 2017, 29, 1605604. [Google Scholar] [CrossRef]
- Ren, E.; Chu, C.; Zhang, Y.; Wang, J.; Pang, X.; Lin, X.; Liu, C.; Shi, X.; Dai, Q.; Lv, P.; et al. Mimovirus Vesicle-Based Biological Orthogonal Reaction for Cancer Diagnosis. Small Methods 2020, 4, 2000291. [Google Scholar] [CrossRef]
- Traïkia, M.; Warschawski, D.E.; Recouvreur, M.; Cartaud, J.; Devaux, P.F. Formation of unilamellar vesicles by repetitive freeze-thaw cycles: Characterization by electron microscopy and 31P-nuclear magnetic resonance. Eur. Biophys. J. 2000, 29, 184–195. [Google Scholar] [CrossRef]
- Lee, D.K.; Kwon, B.S.; Ramamoorthy, A. Freezing point depression of water in phospholipid membranes: A solid-state NMR study. Langmuir 2008, 24, 13598–13604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doux Jacques, P.F.; Hall Benjamin, A.; Killian, J.A. How Lipid Headgroups Sense the Membrane Environment: An Application of 14N NMR. Biophys. J. 2012, 103, 1245–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.-a.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Zhang, X.; Tang, J.; Lv, Q.; Liu, J. Gene-engineered exosomes-thermosensitive liposomes hybrid nanovesicles by the blockade of CD47 signal for combined photothermal therapy and cancer immunotherapy. Biomaterials 2021, 275, 120964. [Google Scholar] [CrossRef] [PubMed]
- Doskocz, J.; Dałek, P.; Przybyło, M.; Trzebicka, B.; Foryś, A.; Kobyliukh, A.; Iglič, A.; Langner, M. The Elucidation of the Molecular Mechanism of the Extrusion Process. Materials 2021, 14, 4278. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.G.M.; Chitneni, M.; Lee, K.S.; Ming, L.C.; Yuen, K.H. Evaluation of Extrusion Technique for Nanosizing Liposomes. Pharmaceutics 2016, 8, 36. [Google Scholar] [CrossRef]
- Jhan, Y.-Y.; Prasca-Chamorro, D.; Palou Zuniga, G.; Moore, D.M.; Arun Kumar, S.; Gaharwar, A.K.; Bishop, C.J. Engineered extracellular vesicles with synthetic lipids via membrane fusion to establish efficient gene delivery. Int. J. Pharm. 2020, 573, 118802. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.J.W.; van de Wakker, S.I.; de Groot, E.M.; de Jong, O.G.; Gitz-François, J.J.J.; Seinen, C.S.; Sluijter, J.P.G.; Schiffelers, R.M.; Vader, P. Functional siRNA Delivery by Extracellular Vesicle–Liposome Hybrid Nanoparticles. Adv. Healthc. Mater. 2022, 11, 2101202. [Google Scholar] [CrossRef] [PubMed]
- Lentz, B.R.; Lee, J.K. Poly(ethylene glycol) (PEG)-mediated fusion between pure lipid bilayers: A mechanism in common with viral fusion and secretory vesicle release? Mol. Membr. Biol. 1999, 16, 279–296. [Google Scholar] [CrossRef]
- Yoshihara, A.; Watanabe, S.; Goel, I.; Ishihara, K.; Ekdahl, K.N.; Nilsson, B.; Teramura, Y. Promotion of cell membrane fusion by cell-cell attachment through cell surface modification with functional peptide-PEG-lipids. Biomaterials 2020, 253, 120113. [Google Scholar] [CrossRef]
- Piffoux, M.; Silva, A.K.A.; Wilhelm, C.; Gazeau, F.; Tareste, D. Modification of Extracellular Vesicles by Fusion with Liposomes for the Design of Personalized Biogenic Drug Delivery Systems. ACS Nano 2018, 12, 6830–6842. [Google Scholar] [CrossRef]
- Kannavou, M.; Marazioti, A.; Stathopoulos, G.T.; Antimisiaris, S.G. Engineered versus hybrid cellular vesicles as efficient drug delivery systems: A comparative study with brain targeted vesicles. Drug Deliv. Transl. Res. 2021, 11, 547–565. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [Green Version]
- Elzanowska, J.; Semira, C.; Costa-Silva, B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2021, 15, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Luo, Z.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Jin, Z.; Manda, K.A.; Isaac, R.; Yang, M.; Fu, W.; et al. Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nat. Metab. 2021, 3, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell Vesicles 2018, 7, 1440131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.; Pazos, R.; Royo, F.; González, E.; Roura-Ferrer, M.; Martinez, A.; Gamiz, J.; Reichardt, N.-C.; Falcón-Pérez, J.M. Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep. 2019, 9, 11920. [Google Scholar] [CrossRef] [Green Version]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The Intracellular Interactome of Tetraspanin-enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes*. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [Green Version]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar] [CrossRef]
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019, 17, 337–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ståhl, A.L.; Johansson, K.; Mossberg, M.; Kahn, R.; Karpman, D. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr. Nephrol. 2019, 34, 11–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Li, S.; Yi, M.; Li, N.; Wu, K. Roles of Microvesicles in Tumor Progression and Clinical Applications. Int. J. Nanomed. 2021, 16, 7071–7090. [Google Scholar] [CrossRef] [PubMed]
- Bagi, Z.; Couch, Y.; Broskova, Z.; Perez-Balderas, F.; Yeo, T.; Davis, S.; Fischer, R.; Sibson, N.R.; Davis, B.G.; Anthony, D.C. Extracellular vesicle integrins act as a nexus for platelet adhesion in cerebral microvessels. Sci. Rep. 2019, 9, 15847. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Wang, X.; Yang, H.; Li, J.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xing, J.; Dai, Z.; Wang, D.; Tang, D. Exosomes: The key of sophisticated cell–cell communication and targeted metastasis in pancreatic cancer. Cell Commun. Signal. 2022, 20, 9. [Google Scholar] [CrossRef]
- Bang, C.; Thum, T. Exosomes: New players in cell–cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef]
- Tian, Z.; Gong, J.; Crowe, M.; Lei, M.; Li, D.; Ji, B.; Diao, J. Biochemical studies of membrane fusion at the single-particle level. Prog. Lipid Res. 2019, 73, 92–100. [Google Scholar] [CrossRef]
- Joardar, A.; Pattnaik, G.P.; Chakraborty, H. Mechanism of Membrane Fusion: Interplay of Lipid and Peptide. J. Membr. Biol. 2022, 255, 211–224. [Google Scholar] [CrossRef]
- Dyer, R.B.; Eller, M.W. Dynamics of Hemagglutinin-Mediated Membrane Fusion. PNAS 2018, 115, 8655–8657. Available online: https://www.pnas.org/doi/abs/10.1073/pnas.1811183115 (accessed on 20 August 2018). [CrossRef] [Green Version]
- Morandi, M.I.; Busko, P.; Ozer-Partuk, E.; Khan, S.; Zarfati, G.; Elbaz-Alon, Y.; Abou Karam, P.; Napso Shogan, T.; Ginini, L.; Gil, Z.; et al. Extracellular vesicle fusion visualized by cryo-electron microscopy. PNAS Nexus 2022, 1, pgac156. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Huo, P.; Liu, B. Formulation Strategies for Folate-Targeted Liposomes and Their Biomedical Applications. Pharmaceutics 2019, 11, 381. [Google Scholar] [CrossRef] [Green Version]
- Olusanya, T.O.B.; Haj Ahmad, R.R.; Ibegbu, D.M.; Smith, J.R.; Elkordy, A.A. Liposomal Drug Delivery Systems and Anticancer Drugs. Molecules 2018, 23, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Joyce, P.; Bremmell, K.; Milne, R.; Prestidge, C.A. Liposomal 5-Fluorouracil Polymer Complexes Facilitate Tumor-Specific Delivery: Pharmaco-Distribution Kinetics Using Microdialysis. Pharmaceutics 2022, 14, 221. [Google Scholar] [CrossRef] [PubMed]
- Nishio, M.; Teranishi, Y.; Morioka, K.; Yanagida, A.; Shoji, A. Real-time assay for exosome membrane fusion with an artificial lipid membrane based on enhancement of gramicidin A channel conductance. Biosens. Bioelectron. 2020, 150, 111918. [Google Scholar] [CrossRef]
- Wu, Q.; Fu, S.; Xiao, H.; Du, J.; Cheng, F.; Wan, S.; Zhu, H.; Li, D.; Peng, F.; Ding, X.; et al. Advances in Extracellular Vesicle Nanotechnology for Precision Theranostics. Adv. Sci. 2023, 10, 2204814. [Google Scholar] [CrossRef]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Ramasubramanian, L.; Du, S.; Gidda, S.; Bahatyrevich, N.; Hao, D.; Kumar, P.; Wang, A. Bioengineering Extracellular Vesicles for the Treatment of Cardiovascular Diseases. Adv. Biol. 2022, 6, 2200087. [Google Scholar] [CrossRef] [PubMed]
- Pick, H.; Alves, A.C.; Vogel, H. Single-Vesicle Assays Using Liposomes and Cell-Derived Vesicles: From Modeling Complex Membrane Processes to Synthetic Biology and Biomedical Applications. Chem. Rev. 2018, 118, 8598–8654. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Iqbal, Z.; Lu, J.; Wang, J.; Zhang, H.; Chen, X.; Duan, L.; Xia, J. Cell-derived nanovesicle-mediated drug delivery to the brain: Principles and strategies for vesicle engineering. Mol. Ther. 2022, 31, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-p.; Lin, Z.-x.; Jiang, X.-y.; Yu, X.-y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, Y.; Akiyoshi, K. Design and function of smart biomembrane nanohybrids for biomedical applications: Review. Polym. J. 2021, 53, 587–592. [Google Scholar] [CrossRef]
- Rayamajhi, S.; Nguyen, T.D.T.; Marasini, R.; Aryal, S. Macrophage-derived exosome-mimetic hybrid vesicles for tumor targeted drug delivery. Acta Biomater. 2019, 94, 482–494. [Google Scholar] [CrossRef]
- Kumar, S.; Karmacharya, M.; Michael, I.J.; Choi, Y.; Kim, J.; Kim, I.; Cho, Y.-K. Programmed exosome fusion for energy generation in living cells. Nat. Catal. 2021, 4, 763–774. [Google Scholar] [CrossRef]
- Liu, H.; Huang, L.; Mao, M.; Ding, J.; Wu, G.; Fan, W.; Yang, T.; Zhang, M.; Huang, Y.; Xie, H.-Y. Viral Protein-Pseudotyped and siRNA-Electroporated Extracellular Vesicles for Cancer Immunotherapy. Adv. Funct. Mater. 2020, 30, 2006515. [Google Scholar] [CrossRef]
- Nguyen, V.-N.; Dao, T.N.T.; Cho, M.; Jeong, H.; Nguyen-Le, M.-T.; Shin, Y.; Yoon, J. Recent advances in extracellular vesicle-based organic nanotherapeutic drugs for precision cancer therapy. Coord. Chem. Rev. 2023, 479, 215006. [Google Scholar] [CrossRef]
- Tang, J.; Su, T.; Huang, K.; Dinh, P.-U.; Wang, Z.; Vandergriff, A.; Hensley, M.T.; Cores, J.; Allen, T.; Li, T.; et al. Targeted repair of heart injury by stem cells fused with platelet nanovesicles. Nat. Biomed. Eng. 2018, 2, 17–26. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome Uptake through Clathrin-mediated Endocytosis and Macropinocytosis and Mediating miR-21 Delivery*. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.; Ughetto, S.; Mahjoum, S.; Nair, A.V.; Breakefield, X.O. Uptake, functionality, and re-release of extracellular vesicle-encapsulated cargo. Cell Rep. 2022, 39, 110651. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. Available online: https://www.mdpi.com/1422-0067/21/20/7568 (accessed on 14 October 2020). [CrossRef]
- Michael, I.; Kim, D.; Gulenko, O.; Kumar, S.; Kumar, S.; Clara, J.; Ki, D.Y.; Park, J.; Jeong, H.Y.; Kim, T.S.; et al. A fidget spinner for the point-of-care diagnosis of urinary tract infection. Nat. Biomed. Eng. 2020, 4, 591–600. [Google Scholar] [CrossRef]
- Leone, D.A.; Rees, A.J.; Kain, R. Dendritic cells and routing cargo into exosomes. Immunol. Cell Biol. 2018, 96, 683–693. [Google Scholar] [CrossRef] [Green Version]
- Glebov, O.O.; Bright, N.A.; Nichols, B.J. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 2006, 8, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shi, H.; Chen, X.; Wang, Z. Regulation of EGF-Stimulated EGF Receptor Endocytosis During M Phase. Traffic 2011, 12, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, H.; Tanowitz, M.; Liang, X.-h.; Crooke, S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 7314–7330. [Google Scholar] [CrossRef] [Green Version]
- Nigri, J.; Leca, J.; Tubiana, S.-S.; Finetti, P.; Guillaumond, F.; Martinez, S.; Lac, S.; Iovanna, J.L.; Audebert, S.; Camoin, L.; et al. CD9 Mediates the Uptake of Extracellular Vesicles from Cancer-Associated Fibroblasts that Promote Pancreatic Cancer Cell Aggressiveness. Sci. Signal. 2022, 15, eabg8191. Available online: https://www.science.org/doi/abs/10.1126/scisignal.abg8191 (accessed on 2 August 2022). [CrossRef]
- Li, Q.; Song, Y.; Wang, Q.; Chen, J.; Gao, J.; Tan, H.; Li, S.; Wu, Y.; Yang, H.; Huang, H.; et al. Engineering extracellular vesicles with platelet membranes fusion enhanced targeted therapeutic angiogenesis in a mouse model of myocardial ischemia reperfusion. Theranostics 2021, 11, 3916–3931. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal. Transduct. Target Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.-S.; Chen, C.-A.; Zhou, Q.A. Exosomes─Nature’s Lipid Nanoparticles, a Rising Star in Drug Delivery and Diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Huang, Z.; Zhong, H.; Lei, Q.; Ai, Y.; Xie, Z.; Zhang, T.; Jiang, B.; Zhu, W.; Sheng, Y.; et al. Analysis and Biomedical Applications of Functional Cargo in Extracellular Vesicles. ACS Nano 2022, 16, 19980–20001. [Google Scholar] [CrossRef]
- Peshkova, M.; Kosheleva, N.; Shpichka, A.; Radenska-Lopovok, S.; Telyshev, D.; Lychagin, A.; Li, F.; Timashev, P.; Liang, X.-J. Targeting Inflammation and Regeneration: Scaffolds, Extracellular Vesicles, and Nanotechnologies as Cell-Free Dual-Target Therapeutic Strategies. Int. J. Mol. Sci. 2022, 23, 13796. [Google Scholar] [CrossRef]
- Sunkara, V.; Kumar, S.; Sabaté del Río, J.; Kim, I.; Cho, Y.-K. Lab-on-a-Disc for Point-of-Care Infection Diagnostics. Acc. Chem. Res. 2021, 54, 3643–3655. [Google Scholar] [CrossRef]
- Kumar, S.; Rhim, W.-K.; Lim, D.-K.; Nam, J.-M. Glutathione Dimerization-Based Plasmonic Nanoswitch for Biodetection of Reactive Oxygen and Nitrogen Species. ACS Nano 2013, 7, 2221–2230. [Google Scholar] [CrossRef]
- Kumar, S.; Han, J.-A.; Michael, I.J.; Ki, D.; Sunkara, V.; Park, J.; Gautam, S.; Ha, H.K.; Zhang, L.; Cho, Y.-K. Human Platelet Membrane Functionalized Microchips with Plasmonic Codes for Cancer Detection. Adv. Funct. Mater. 2019, 29, 1902669. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, A.; Kim, G.-H.; Rhim, W.-K.; Hartman, K.L.; Nam, J.-M. Myoglobin and Polydopamine-Engineered Raman Nanoprobes for Detecting, Imaging, and Monitoring Reactive Oxygen Species in Biological Samples and Living Cells. Nano Micro Small 2017, 13, 1701584. [Google Scholar] [CrossRef]
- Wang, C.; Yan, A.; Wang, H.; Su, Y.; Li, D. DNA-Mediated Membrane Fusion and Its Biological Applications: Sensing, Reaction Control and Drug Delivery. Anal. Sens. 2022, 2, e202200024. [Google Scholar] [CrossRef]
- Ning, B.; Huang, Z.; Youngquist, B.M.; Scott, J.W.; Niu, A.; Bojanowski, C.M.; Zwezdaryk, K.J.; Saba, N.S.; Fan, J.; Yin, X.-M.; et al. Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma. Nat. Nanotechnol. 2021, 16, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Jumeaux, C.; Wahlsten, O.; Block, S.; Kim, E.; Chandrawati, R.; Howes, P.D.; Höök, F.; Stevens, M.M. MicroRNA Detection by DNA-Mediated Liposome Fusion. Chembiochem 2018, 19, 434–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Li, S.; Ding, F.; Fan, H.; Shi, L.; Zhu, L.; Li, J.; Feng, J.; Zhu, X.; Zhang, C. Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew Chem. Int. Ed. Engl. 2019, 58, 8719–8723. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Ho, E.A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Control Release 2021, 329, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.; Yi, Y.; Yu, Y. Effect of partial PEGylation on particle uptake by macrophages. Nanoscale 2017, 9, 288–297. [Google Scholar] [CrossRef]
- Zhang, N.; Song, Y.; Huang, Z.; Chen, J.; Tan, H.; Yang, H.; Fan, M.; Li, Q.; Wang, Q.; Gao, J.; et al. Monocyte mimics improve mesenchymal stem cell-derived extracellular vesicle homing in a mouse MI/RI model. Biomaterials 2020, 255, 120168. [Google Scholar] [CrossRef]
- Chakravarti, A.R.; Pacelli, S.; Paul, A. Investigation of human adipose stem cell-derived nanoparticles as a biomimetic carrier for intracellular drug delivery. Nanoscale 2020, 12, 24273–24284. [Google Scholar] [CrossRef]
- Wan, Y.; Wang, L.; Zhu, C.; Zheng, Q.; Wang, G.; Tong, J.; Fang, Y.; Xia, Y.; Cheng, G.; He, X.; et al. Aptamer-Conjugated Extracellular Nanovesicles for Targeted Drug Delivery. Cancer Res. 2018, 78, 798–808. [Google Scholar] [CrossRef] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karmacharya, M.; Kumar, S.; Cho, Y.-K. Tuning the Extracellular Vesicles Membrane through Fusion for Biomedical Applications. J. Funct. Biomater. 2023, 14, 117. https://doi.org/10.3390/jfb14020117
Karmacharya M, Kumar S, Cho Y-K. Tuning the Extracellular Vesicles Membrane through Fusion for Biomedical Applications. Journal of Functional Biomaterials. 2023; 14(2):117. https://doi.org/10.3390/jfb14020117
Chicago/Turabian StyleKarmacharya, Mamata, Sumit Kumar, and Yoon-Kyoung Cho. 2023. "Tuning the Extracellular Vesicles Membrane through Fusion for Biomedical Applications" Journal of Functional Biomaterials 14, no. 2: 117. https://doi.org/10.3390/jfb14020117