Mapping the Most Common Founder Variant in RSPH9 That Causes Primary Ciliary Dyskinesia in Multiple Consanguineous Families of Bedouin Arabs
Abstract
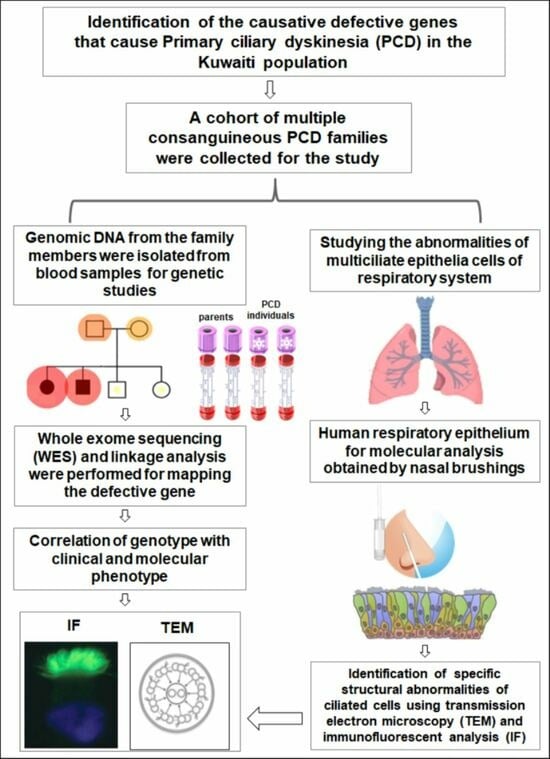
:1. Introduction
Genetics of Primary Ciliary Dyskinesia
2. Methods
2.1. Human Subjects
2.2. Genomic DNA and Exome Sequencing
2.3. Autozygosity Mapping and Variant Screening
2.4. Immunofluorescent Analyses of Nasal Biopsies
2.5. TEM Analysis of Nasal Biopsies
2.6. Validating the RSPH9 Founder Variant in Arab Population
3. Results
3.1. Autozygosity Mapping Reveals a Founder Homozygous Inframe Deletion in RSPH9
3.2. Detection of Compound Heterozygous Variants in RSPH9 in KU-15.IV-2 Individual
3.3. Detection of Ultrastructural Defects of the Cilia
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magnin, M.L.; Cros, P.; Beydon, N.; Mahloul, M.; Tamalet, A.; Escudier, E.; Clement, A.; Le Pointe, H.D.; Blanchon, S. Longitudinal lung function and structural changes in children with primary ciliary dyskinesia. Pediatr. Pulmonol. 2012, 47, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Zariwala, M.A.; Omran, H.; Ferkol, T.W. The emerging genetics of primary ciliary dyskinesia. Proc. Am. Thorac. Soc. 2011, 8, 430–433. [Google Scholar] [CrossRef]
- Hannah, W.B.; Seifert, B.A.; Truty, R.; Zariwala, M.A.; Ameel, K.; Zhao, Y.; Nykamp, K.; Gaston, B. The global prevalence and ethnic heterogeneity of primary ciliary dyskinesia gene variants: A genetic database analysis. Lancet Respir. Med. 2022, 10, 459–468. [Google Scholar] [CrossRef]
- Zariwala, M.A.; Knowles, M.R.; Omran, H. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 2007, 69, 423–450. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.R.; Daniels, L.A.; Davis, S.D.; Zariwala, M.A.; Leigh, M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013, 188, 913–922. [Google Scholar] [CrossRef]
- Horani, A.; Ferkol, T.W.; Dutcher, S.K.; Brody, S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016, 18, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Leigh, M.W.; Horani, A.; Kinghorn, B.; O’Connor, M.G.; Zariwala, M.A.; Knowles, M.R. Primary Ciliary Dyskinesia (PCD): A genetic disorder of motile cilia. Transl. Sci. Rare Dis. 2019, 4, 51–75. [Google Scholar] [CrossRef]
- Ibanez-Tallon, I.; Heintz, N.; Omran, H. To beat or not to beat: Roles of cilia in development and disease. Hum. Mol. Genet. 2003, 12 (Suppl. S1), R27–R35. [Google Scholar] [CrossRef]
- Kott, E.; Legendre, M.; Copin, B.; Papon, J.F.; Dastot-Le Moal, F.; Montantin, G.; Duquesnoy, P.; Piterboth, W.; Amram, D.; Bassinet, L.; et al. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 2013, 93, 561–570. [Google Scholar] [CrossRef]
- Castleman, V.H.; Romio, L.; Chodhari, R.; Hirst, R.A.; de Castro, S.C.P.; Parker, K.A.; Ybot-Gonzalez, P.; Emes, R.D.; Wilson, S.W.; Wallis, C.; et al. Mutations in Radial Spoke Head Protein Genes RSPH9 and RSPH4A Cause Primary Ciliary Dyskinesia with Central-Microtubular-Pair Abnormalities. Am. J. Hum. Genet. 2009, 84, 197–209. [Google Scholar] [CrossRef]
- Jeanson, L.; Copin, B.; Papon, J.F.; Dastot-Le Moal, F.; Duquesnoy, P.; Montantin, G.; Cadranel, J.; Corvol, H.; Coste, A.; Desir, J.; et al. RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am. J. Hum. Genet. 2015, 97, 153–162. [Google Scholar] [CrossRef] [PubMed]
- El Khouri, E.; Thomas, L.; Jeanson, L.; Bequignon, E.; Vallette, B.; Duquesnoy, P.; Montantin, G.; Copin, B.; Dastot-Le Moal, F.; Blanchon, S.; et al. Mutations in DNAJB13, Encoding an HSP40 Family Member, Cause Primary Ciliary Dyskinesia and Male Infertility. Am. J. Hum. Genet. 2016, 99, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.H.; Huh, H.J.; Jeong, I.; Lee, N.Y.; Koh, W.J.; Park, H.C.; Ki, C.S. A nonsense variant in NME5 causes human primary ciliary dyskinesia with radial spoke defects. Clin. Genet. 2020, 98, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Anderegg, L.; Im Hof Gut, M.; Hetzel, U.; Howerth, E.W.; Leuthard, F.; Kyostila, K.; Lohi, H.; Pettitt, L.; Mellersh, C.; Minor, K.M.; et al. NME5 frameshift variant in Alaskan Malamutes with primary ciliary dyskinesia. PLoS Genet. 2019, 15, e1008378. [Google Scholar] [CrossRef] [PubMed]
- Al-Mutairi, D.A.; Alsabah, B.H.; Alkhaledi, B.A.; Pennekamp, P.; Omran, H. Identification of a novel founder variant in DNAI2 cause primary ciliary dyskinesia in five consanguineous families derived from a single tribe descendant of Arabian Peninsula. Front. Genet. 2022, 13, 1017280. [Google Scholar] [CrossRef]
- Watson, C.M.; Crinnion, L.A.; Gurgel-Gianetti, J.; Harrison, S.M.; Daly, C.; Antanavicuite, A.; Lascelles, C.; Markham, A.F.; Pena, S.D.; Bonthron, D.T.; et al. Rapid Detection of Rare Deleterious Variants by Next Generation Sequencing with Optional Microarray SNP Genotype Data. Hum. Mutat. 2015, 36, 823–830. [Google Scholar] [CrossRef]
- Carr, I.M.; Camm, N.; Taylor, G.R.; Charlton, R.; Ellard, S.; Sheridan, E.G.; Markham, A.F.; Bonthron, D.T. GeneScreen: A program for high-throughput mutation detection in DNA sequence electropherograms. J. Med. Genet. 2011, 48, 123–130. [Google Scholar] [CrossRef]
- Wallmeier, J.; Al-Mutairi, D.A.; Chen, C.T.; Loges, N.T.; Pennekamp, P.; Menchen, T.; Ma, L.; Shamseldin, H.E.; Olbrich, H.; Dougherty, G.W.; et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Genet. 2014, 46, 646–651. [Google Scholar] [CrossRef]
- Knowles, M.R.; Ostrowski, L.E.; Leigh, M.W.; Sears, P.R.; Davis, S.D.; Wolf, W.E.; Hazucha, M.J.; Carson, J.L.; Olivier, K.N.; Sagel, S.D.; et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 2014, 189, 707–717. [Google Scholar] [CrossRef]
- Carr, I.M.; Bhaskar, S.; O’ Sullivan, J.; Aldahmesh, M.A.; Shamseldin, H.E.; Markham, A.F.; Bonthron, D.T.; Black, G.; Alkuraya, F.S. Autozygosity Mapping with Exome Sequence Data. Hum. Mutat. 2012, 34, 50–56. [Google Scholar] [CrossRef]
- Carr, I.M.; Morgan, J.; Watson, C.; Melnik, S.; Diggle, C.P.; Logan, C.V.; Harrison, S.M.; Taylor, G.R.; Pena, S.D.; Markham, A.F.; et al. Simple and efficient identification of rare recessive pathologically important sequence variants from next generation exome sequence data. Hum. Mutat. 2013, 34, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Niziolek, M.; Bicka, M.; Osinka, A.; Samsel, Z.; Sekretarska, J.; Poprzeczko, M.; Bazan, R.; Fabczak, H.; Joachimiak, E.; Wloga, D. PCD Genes-From Patients to Model Organisms and Back to Humans. Int. J. Mol. Sci. 2022, 23, 1749. [Google Scholar] [CrossRef] [PubMed]
- Reish, O.; Slatkin, M.; Chapman-Shimshoni, D.; Elizur, A.; Chioza, B.; Castleman, V.; Mitchison, H.M. Founder mutation(s) in the RSPH9 gene leading to primary ciliary dyskinesia in two inbred Bedouin families. Ann. Hum. Genet. 2010, 74, 117–125. [Google Scholar] [CrossRef]
- Alsaadi, M.M.; Gaunt, T.R.; Boustred, C.R.; Guthrie, P.A.; Liu, X.; Lenzi, L.; Rainbow, L.; Hall, N.; Alharbi, K.K.; Day, I.N. From a single whole exome read to notions of clinical screening: Primary ciliary dyskinesia and RSPH9 p.Lys268del in the Arabian Peninsula. Ann. Hum. Genet. 2012, 76, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Botstein, D. Homozygosity mapping: A way to map human recessive traits with the DNA of inbred children. Science 1987, 236, 1567–1570. [Google Scholar] [CrossRef]
- Panizzi, J.R.; Becker-Heck, A.; Castleman, V.H.; Al-Mutairi, D.A.; Liu, Y.; Loges, N.T.; Pathak, N.; Austin-Tse, C.; Sheridan, E.; Schmidts, M.; et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012, 44, 714–719. [Google Scholar] [CrossRef]
- Zariwala, M.A.; Gee, H.Y.; Kurkowiak, M.; Al-Mutairi, D.A.; Leigh, M.W.; Hurd, T.W.; Hjeij, R.; Dell, S.D.; Chaki, M.; Dougherty, G.W.; et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013, 93, 336–345. [Google Scholar] [CrossRef]
- Fassad, M.R.; Shoman, W.I.; Morsy, H.; Patel, M.P.; Radwan, N.; Jenkins, L.; Cullup, T.; Fouda, E.; Mitchison, H.M.; Fasseeh, N. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin. Genet. 2020, 97, 509–515. [Google Scholar] [CrossRef]
- Monies, D.; Abouelhoda, M.; Assoum, M.; Moghrabi, N.; Rafiullah, R.; Almontashiri, N.; Alowain, M.; Alzaidan, H.; Alsayed, M.; Subhani, S.; et al. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019, 104, 1182–1201. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Frommer, A.; Hjeij, R.; Loges, N.T.; Edelbusch, C.; Jahnke, C.; Raidt, J.; Werner, C.; Wallmeier, J.; Grosse-Onnebrink, J.; Olbrich, H.; et al. Immunofluorescence Analysis and Diagnosis of Primary Ciliary Dyskinesia with Radial Spoke Defects. Am. J. Respir. Cell Mol. Biol. 2015, 53, 563–573. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Mutairi, D.A.; Alsabah, B.H.; Pennekamp, P.; Omran, H. Mapping the Most Common Founder Variant in RSPH9 That Causes Primary Ciliary Dyskinesia in Multiple Consanguineous Families of Bedouin Arabs. J. Clin. Med. 2023, 12, 6505. https://doi.org/10.3390/jcm12206505
Al-Mutairi DA, Alsabah BH, Pennekamp P, Omran H. Mapping the Most Common Founder Variant in RSPH9 That Causes Primary Ciliary Dyskinesia in Multiple Consanguineous Families of Bedouin Arabs. Journal of Clinical Medicine. 2023; 12(20):6505. https://doi.org/10.3390/jcm12206505
Chicago/Turabian StyleAl-Mutairi, Dalal A., Basel H. Alsabah, Petra Pennekamp, and Heymut Omran. 2023. "Mapping the Most Common Founder Variant in RSPH9 That Causes Primary Ciliary Dyskinesia in Multiple Consanguineous Families of Bedouin Arabs" Journal of Clinical Medicine 12, no. 20: 6505. https://doi.org/10.3390/jcm12206505