Reinfection with SARS-CoV-2 and Waning Humoral Immunity: A Case Report
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Population & PCR Testing
2.2. Virologic and Immunologic Analyses
3. Results
3.1. Population Sampling
3.2. Case Study
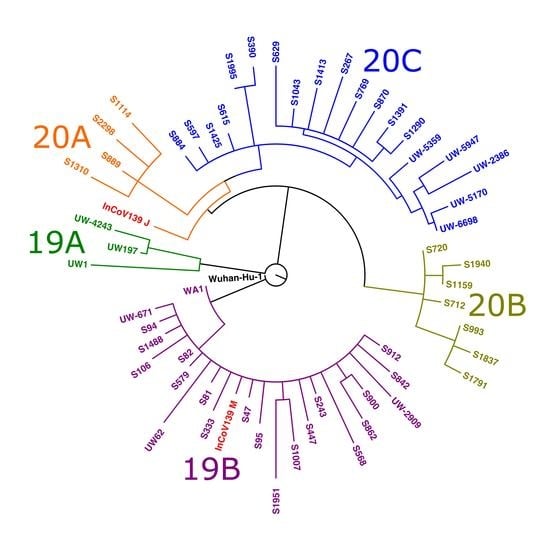
3.3. Viral Sequencing and Phylogenetic Analysis
3.4. Anti-SARS-CoV-2 Antibody Response
3.5. Antibody and B-Cell Receptor Repertoires
3.6. T Cell Phenotypes and Plasma Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Duration of Isolation and Precautions for Adults with COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html (accessed on 26 August 2020).
- Xu, K.; Chen, Y.; Yuan, J.; Yi, P.; Ding, C.; Wu, W.; Li, Y.; Ni, Q.; Zou, R.; Li, X.; et al. Factors Associated With Prolonged Viral RNA Shedding in Patients with Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2020, 71, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arons, M.M.; Hatfield, K.M.; Reddy, S.C.; Kimball, A.; James, A.; Jacobs, J.R.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N. Engl. J. Med. 2020, 382, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Van Kampen, J.J.A.; Van De Vijver, D.A.M.C.; Fraaij, P.L.A.; Haagmans, B.L.; Lamers, M.M.; Okba, N.; van den Akker, J.P.C.; Endeman, H.; Gommers, D.A.M.P.J.; Cornelissen, J.J.; et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat. Commun. 2021, 12, 267. [Google Scholar] [CrossRef]
- KCDC. Division of Risk Assessment and International Cooperation, Korean Center for Disease Control. Findings from Investigation and Analysis of Re-Positive Cases. Available online: https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030&act=view&list_no=367267&nPage=1 (accessed on 26 August 2020).
- Wang, P. Recurrent presence of SARS-CoV-2 RNA in a 33-year-old man. J. Med. Virol. 2020, 93, 592–594. [Google Scholar] [CrossRef]
- Duggan, N.M.; Ludy, S.M.; Shannon, B.C.; Reisner, A.T.; Wilcox, S.R. Is novel coronavirus 2019 reinfection possible? Interpreting dynamic SARS-CoV-2 test results. Am. J. Emerg. Med. 2020, 39, 256.e1–256.e3. [Google Scholar] [CrossRef]
- Yuan, B.; Liu, H.-Q.; Yang, Z.-R.; Chen, Y.-X.; Liu, Z.-Y.; Zhang, K.; Wang, C.; Li, W.-X.; An, Y.-W.; Wang, J.-C.; et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci. Rep. 2020, 10, 11887. [Google Scholar] [CrossRef]
- To, K.K.; Hung, I.F.; Ip, J.D.; Chu, A.W.; Chan, W.M.; Tam, A.R.; Fong, C.H.; Yuan, S.; Tsoi, H.W.; Ng, A.C.; et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2020, 73, e2946–e2951. [Google Scholar] [CrossRef]
- Tillett, R.L.; Sevinsky, J.R.; Hartley, P.D.; Kerwin, H.; Crawford, N.; Gorzalski, A.; Laverdure, C.; Verma, S.C.; Rossetto, C.C.; Jackson, D.; et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021, 21, 52–58. [Google Scholar] [CrossRef]
- Whitman, J.D.; Hiatt, J.; Mowery, C.T.; Shy, B.R.; Yu, R.; Yamamoto, T.N.; Rathore, U.; Goldgof, G.M.; Whitty, C.; Woo, J.M.; et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv 2020. [Google Scholar] [CrossRef]
- Röltgen, K.; Wirz, O.F.; Stevens, B.A.; Powell, A.E.; Hogan, C.A.; Najeeb, J.; Hunter, M.; Sahoo, M.K.; Huang, C.; Yamamoto, F.; et al. SARS-CoV-2 Antibody Responses Correlate with Resolution of RNAemia But Are Short-Lived in Patients with Mild Illness. medRxiv 2020. [Google Scholar] [CrossRef]
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.-L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, e02107-20. [Google Scholar] [CrossRef] [PubMed]
- Joyner, M.J.; Senefeld, J.W.; Klassen, S.A.; John, R.M.; Johnson, P.W.; Theel, E.S.; Wiggins, C.C.; Bruno, K.A.; Klompas, A.M.; Lesser, E.R.; et al. Effect of convalescent plasma on mortality among hospitalized patients with COVID-19: Initial three-month experience. medRxiv 2020. [Google Scholar] [CrossRef]
- Corey, B.L.; Mascola, J.R.; Fauci, A.S.; Collins, F.S. A strategic approach to COVID-19 vaccine R & D. Science 2020, 368, 948–950. [Google Scholar] [CrossRef] [PubMed]
- CDC. Discontinuation of Transmission-Based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings (Interim Guidance). Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html (accessed on 26 August 2020).
- Bedford, T.; Greninger, A.L.; Roychoudhury, P.; Starita, L.M.; Famulare, M.; Huang, M.-L.; Nalla, A.; Pepper, G.; Reinhardt, A.; Xie, H.; et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science 2020, 370, 571–575. [Google Scholar] [CrossRef]
- Quick, J. nCoV-2019 Sequencing Protocol v3. Available online: https://github.com/joshquick/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/nCoV-2019.tsv (accessed on 22 July 2020).
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [Green Version]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Nielsen, S.C.A.; Yang, F.; Jackson, K.J.L. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020, 28, 516–525. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Lausted, C. Multiomic Immunophenotyping of COVID-19 Patients Reveals Early Infection Trajectories. bioRxiv 2020. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Wagner, C.; Vaughan, T.; Stadler, T.; Bedford, T. Analyses of the hCoV-19 outbreak in Washington State: Phylodynamic Analysis of COVID-19 Outbreak in Washington State Shows Rapid Early Growth Followed by Substantial Reductions in Transmission. Available online: https://bedford.io/projects/ncov-wa-phylodynamics/ (accessed on 29 August 2020).
- Goldman, J.D.; Wang, K.; Roltgen, K.; Nielsen, S.C.A.; Roach, J.C.; Naccache, S.N.; Yang, F.; Wirz, O.F.; Yost, K.E.; Lee, J.-Y.; et al. Reinfection with SARS-CoV-2 and Failure of Humoral Immunity: A case report. medRxiv 2020. [Google Scholar] [CrossRef]
- Alvarez-Moreno, C.A.; Rodríguez-Morales, A.J. Testing Dilemmas: Post negative, positive SARS-CoV-2 RT-PCR—Is it a reinfection? Travel Med. Infect. Dis. 2020, 35, 101743. [Google Scholar] [CrossRef] [PubMed]
- Hethcote, H.W. Three Basic Epidemiological Models; Springer: Berlin/Heidelberg, Germany, 1989; pp. 119–144. [Google Scholar] [CrossRef]
- Ferrante, L.; Duczmal, L.H.; Steinmetz, W.A.; Almeida, A.C.L.; Leão, J.; Vassão, R.C.; Tupinambás, U.; Fearnside, P.M. Brazil’s COVID-19 Epicenter in Manaus: How Much of the Population Has Already Been Exposed and Are Vulnerable to SARS-CoV-2? J. Racial Ethn. Health Disparities 2021, 9, 2098–2104. [Google Scholar] [CrossRef] [PubMed]
- Chamblain, M.; Dawkins, E.; Lane, J.; Ghany, R.; Tamariz, L.; Palacio, A.; Guzman-Suarez, B. A case of COVID-19 Reinfection and Systematic Review of Patterns of Reinfection. Infect. Dis. Clin. Pract. 2021, 29, e409–e411. [Google Scholar] [CrossRef]
- Flacco, M.E.; Martellucci, C.A.; Baccolini, V.; De Vito, C.; Renzi, E.; Villari, P.; Manzoli, L. Risk of reinfection and disease after SARS-CoV-2 primary infection: Meta-analysis. Eur. J. Clin. Investig. 2022, 52, e13845. [Google Scholar] [CrossRef]
- Nalla, A.K.; Casto, A.M.; Huang, M.-L.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2020, 58, e00557-20. [Google Scholar] [CrossRef] [Green Version]
- Lieberman, J.A.; Pepper, G.; Naccache, S.N.; Huang, M.-L.; Jerome, K.R.; Greninger, A.L. Comparison of Commercially Available and Laboratory-Developed Assays for In Vitro Detection of SARS-CoV-2 in Clinical Laboratories. J. Clin. Microbiol. 2020, 58, e00821-20. [Google Scholar] [CrossRef]
- WHO. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RTPCR-Protocol and Preliminary Evaluation as of Jan 13, 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902.pdf?sfvrsn=d381fc88_2 (accessed on 22 June 2020).
- CDC. CDC’s Diagnostic Test for COVID-19 Only and Supplies. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 4 September 2020).
- Greninger, A.L.; Zerr, D.M.; Qin, X.; Adler, A.L.; Sampoleo, R.; Kuypers, J.M.; Englund, J.A.; Jerome, K.R. Rapid Metagenomic Next-Generation Sequencing during an Investigation of Hospital-Acquired Human Parainfluenza Virus 3 Infections. J. Clin. Microbiol. 2017, 55, 177–182. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef] [Green Version]
- Hodcroft, E.; Hadfield, J.; Neher, R.; Bedford, T. Year-letter Genetic Clade Naming for SARS-CoV-2 on Nextstain.org. Available online: https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming (accessed on 29 August 2020).
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roskin, K.M.; Jackson, K.J.L.; Lee, J.-Y.; Hoh, R.A.; Joshi, S.A.; Hwang, K.-K.; Bonsignori, M.; Pedroza-Pacheco, I.; Liao, H.-X.; Moody, M.A.; et al. Aberrant B cell repertoire selection associated with HIV neutralizing antibody breadth. Nat. Immunol. 2020, 21, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019, 8, 281–291.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef] [Green Version]
- Manor, O.; Zubair, N.; Conomos, M.P.; Xu, X.; Rohwer, J.E.; Krafft, C.E.; Lovejoy, J.C.; Magis, A.T. A Multi-omic Association Study of Trimethylamine N-Oxide. Cell Rep. 2018, 24, 935–946. [Google Scholar] [CrossRef]





| Parameter: | Primary Infection (March) * | Reinfection (July) * |
|---|---|---|
| Vital Signs: | ||
| Temperature (°C) | 38.4 | 37.0 |
| Heart Rate (/min) | 101 | 86 |
| Blood pressure (mmHg) | 156/96 | 143/93 |
| Respiratory Rate (/min) | 20 | 19 |
| SpO2 (%) on supplemental O2 rate | 93% on 6 L/min | 94–97% on 3 L/min |
| BMI (kg/m2) | 18.7 | 20.4 |
| Laboratory: | ||
| Total white blood count (cells/µL) | 16,200 | 6700 |
| Absolute neutrophil count (cells/µL) | 12,960 | 2010 |
| Absolute lymphocyte count (cells/µL) | 1600 | 600 |
| Hematocrit (%) | 39.6% | 42.8% |
| Platelet count (cells/µL) | 290,000 | 240,000 |
| D-dimer (≤0.49 µg/mL) ** | N/A | 0.47 |
| Creatinine (mg/dL) | 1.01 | 1.07 |
| Procalcitonin (≤0.25 ng/mL) ** | 0.15 | 0.08 |
| C-reactive protein (≤5 mg/L) ** | N/A | <3.0 |
| SARS-CoV-2 rt-PCR CT (target 1) † | 22.8 (E) | 43.3 (E) |
| SARS-CoV-2 rt-PCR CT (target 2) † | 26.5 (RdRp) | 39.6 (N2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goldman, J.D.; Wang, K.; Röltgen, K.; Nielsen, S.C.A.; Roach, J.C.; Naccache, S.N.; Yang, F.; Wirz, O.F.; Yost, K.E.; Lee, J.-Y.; et al. Reinfection with SARS-CoV-2 and Waning Humoral Immunity: A Case Report. Vaccines 2023, 11, 5. https://doi.org/10.3390/vaccines11010005
Goldman JD, Wang K, Röltgen K, Nielsen SCA, Roach JC, Naccache SN, Yang F, Wirz OF, Yost KE, Lee J-Y, et al. Reinfection with SARS-CoV-2 and Waning Humoral Immunity: A Case Report. Vaccines. 2023; 11(1):5. https://doi.org/10.3390/vaccines11010005
Chicago/Turabian StyleGoldman, Jason D., Kai Wang, Katharina Röltgen, Sandra C. A. Nielsen, Jared C. Roach, Samia N. Naccache, Fan Yang, Oliver F. Wirz, Kathryn E. Yost, Ji-Yeun Lee, and et al. 2023. "Reinfection with SARS-CoV-2 and Waning Humoral Immunity: A Case Report" Vaccines 11, no. 1: 5. https://doi.org/10.3390/vaccines11010005