Microbial Production of Retinyl Palmitate and Its Application as a Cosmeceutical
Abstract
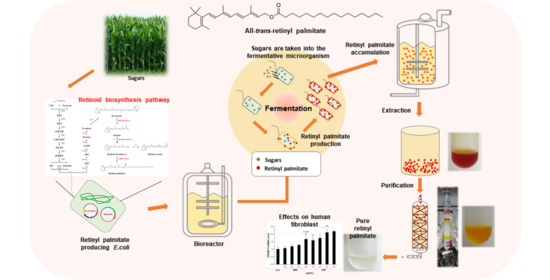
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Plasmid Construction for the Expression of Retinoid Biosynthesis Pathway Enzymes
2.3. Genome Engineering for Increasing Isopentenyl Diphosphate Pools
2.4. Extraction and Analysis of Retinoids
2.5. Batch and Fed-Batch Fermentation
2.6. Cell Viability Assay and Enzyme-Linked Immunosorbent Assay
2.7. Reverse-Transcription PCR and SDS-PAGE Analyses
2.8. Statistical Analysis
3. Results and Discussion
3.1. Construction of the β-Carotene Pathway for Retinol Production
3.2. Retinol Production Using the Expression of Heterologous β-Carotene 15,15′-Oxygenase
3.3. mRNA-Stabilizing Region Engineering of Blh for Enhanced Retinol Production
3.4. Construction of a Retinyl Palmitate Biosynthesis Pathway
3.5. Batch and Fed-Batch Fermentation Procedures for Retinyl Palmitate Production
3.6. Anti-Ageing-Related Effect of Retinyl Palmitate (Obtained from a Microorganism) on Human Skin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blomhoff, R.; Blomhoff, H.K. Overview of retinoid metabolism and function. J. Neurobiol. 2006, 66, 606–630. [Google Scholar] [CrossRef] [PubMed]
- Tanumihardjo, S.A. Vitamin A: Biomarkers of nutrition for development. Am. J. Clin. Nutr. 2011, 94, 658S–665S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, S.; Bardawil, T.; Stephan, C.; Darwiche, N.; Abbas, O.; Kibbi, A.G.; Nemer, G.; Kurban, M. Retinoids: A journey from the molecular structures and mechanisms of action to clinical uses in dermatology and adverse effects. J. Dermatolog. Treat. 2017, 28, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Oh, D.K. Substrate specificity of a recombinant chicken beta-carotene 15,15′-monooxygenase that converts beta-carotene into retinal. Biotechnol. Lett. 2009, 31, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Hoog, J.O.; Ostberg, L.J. Mammalian alcohol dehydrogenases--a comparative investigation at gene and protein levels. Chem. Biol. Interact. 2011, 191, 2–7. [Google Scholar] [CrossRef]
- Menozzi, I.; Vallese, F.; Polverini, E.; Folli, C.; Berni, R.; Zanotti, G. Structural and molecular determinants affecting the interaction of retinol with human CRBP1. J. Struct. Biol. 2017, 197, 330–339. [Google Scholar] [CrossRef]
- O’Byrne, S.M.; Blaner, W.S. Retinol and retinyl esters: Biochemistry and physiology. J. Lipid. Res. 2013, 54, 1731–1743. [Google Scholar] [CrossRef] [Green Version]
- Blaner, W.S.; Olson, J.A. Retinol and Retinoic Acid Metabolism. In The Retinoids: Biology, Chemistry, and Medicine, 2nd ed.; Sporn, M.B., Robert, A.B., Goodman, D.S., Eds.; Raven: New York, NY, USA, 1994; pp. 229–256. [Google Scholar]
- Wyss, A.; Wirtz, G.; Woggon, W.; Brugger, R.; Wyss, M.; Friedlein, A.; Bachmann, H.; Hunziker, W. Cloning and expression of beta, beta-carotene 15,15′-dioxygenase. Biochem. Biophys. Res. Commun. 2000, 271, 334–336. [Google Scholar] [CrossRef]
- Yan, W.; Jang, G.F.; Haeseleer, F.; Esumi, N.; Chang, J.; Kerrigan, M.; Campochiaro, M.; Campochiaro, P.; Palczewski, K.; Zack, D.J. Cloning and characterization of a human beta, beta-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics 2001, 72, 193–202. [Google Scholar] [CrossRef]
- Paik, J.; During, A.; Harrison, E.H.; Mendelsohn, C.L.; Lai, K.; Blaner, W.S. Expression and characterization of a murine enzyme able to cleave beta-carotene. The formation of retinoids. J. Biol. Chem. 2001, 276, 32160–32168. [Google Scholar] [CrossRef] [Green Version]
- Sabehi, G.; Loy, A.; Jung, K.H.; Partha, R.; Spudich, J.L.; Isaacson, T.; Hirschberg, J.; Wagner, M.; Beja, O. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005, 3, e273. [Google Scholar] [CrossRef] [PubMed]
- Baliga, N.S.; Bonneau, R.; Facciotti, M.T.; Pan, M.; Glusman, G.; Deutsch, E.W.; Shannon, P.; Chiu, Y.; Weng, R.S.; Gan, R.R.; et al. Genome sequence of Haloarcula marismortui: A halophilic archaeon from the Dead Sea. Genome Res. 2004, 14, 2221–2234. [Google Scholar] [CrossRef] [Green Version]
- Pfeiffer, F.; Schuster, S.C.; Broicher, A.; Falb, M.; Palm, P.; Rodewald, K.; Ruepp, A.; Soppa, J.; Tittor, J.; Oesterhelt, D. Evolution in the laboratory: The genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 2008, 91, 335–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.; Bradley, A.S.; Waldbauer, J.R.; Summons, R.E.; DeLong, E.F. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. USA 2007, 104, 5590–5595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mongodin, E.F.; Nelson, K.E.; Daugherty, S.; Deboy, R.T.; Wister, J.; Khouri, H.; Weidman, J.; Walsh, D.A.; Papke, R.T.; Sanchez Perez, G.; et al. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. USA 2005, 102, 18147–18152. [Google Scholar] [CrossRef] [Green Version]
- Oren, A. Salinibacter: An extremely halophilic bacterium with archaeal properties. FEMS Microbiol. Lett. 2013, 342, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fisher, G.J.; Wang, Z.Q.; Datta, S.C.; Varani, J.; Kang, S.; Voorhees, J.J. Pathophysiology of Premature Skin Aging Induced by Ultraviolet Light. N. Engl. J. Med. 1997, 337, 1419–1428. [Google Scholar] [CrossRef]
- Jang, H.J.; Ha, B.K.; Zhou, J.; Ahn, J.Y.; Yoon, S.H.; Kim, S.W. Selective Retinol Production by Modulating the Composition of Retinoids from Metabolically Engineered E. coli. Biotechnol. Bioeng. 2015, 112, 1604–1612. [Google Scholar] [CrossRef]
- Sun, L.; Kwak, S.; Jin, Y.S. Vitamin A Production by Engineered Saccharomyces cerevisiae from Xylose via Two-Phase in Situ Extraction. ACS Synth. Biol. 2019, 8, 2131–2140. [Google Scholar] [CrossRef]
- Lian, X.; Fang, Y.; Joseph, E.; Wang, Q.; Li, J.; Banerjee, S.; Lollar, C.; Wang, S.; Zhou, H.C. Enzyme–MOF (metal–organic framework) composites. Chem. Soc. Rev. 2017, 46, 3386–3401. [Google Scholar] [CrossRef]
- Ro, J.; Kim, Y.; Kim, H.; Jang, S.B.; Lee, H.J.; Chakma, S.; Jeong, J.H.; Lee, J. Anti-oxidative activity of pectin and its stabilizing effect on retinyl palmitate. Korean J. Physiol. Pharmacol. 2013, 17, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yu, J.; Wang, A.; Zuo, C.; Li, H.; Chen, X.; Pei, X.; Zhang, P. Efficient synthesis of vitamin A palmitate in nonaqueous medium using self-assembled lipase TLL@apatite hybrid nanoflowers by mimetic biomineralization. Green Chem. Lett. Rev. 2018, 11, 476–483. [Google Scholar] [CrossRef] [Green Version]
- Muthusamy, K.; Sridharan, V.; Maheswari, C.U.; Nagarajan, S. Lipase catalyzed synthesis of fluorescent glycolipids: Gelation studies and graphene incorporated self-assembled sheet formation for semiconductor applications. Green Chem. 2016, 18, 3722–3731. [Google Scholar] [CrossRef]
- Kang, H.; Kim, C.; Ji, E.; Ahn, S.; Jung, M.; Hong, Y.; Kim, W.; Lee, E.K. The MicroRNA-551a/MEF2C Axis Regulates the Survival and Sphere Formation of Cancer Cells in Response to 5-Fluorouracil. Mol. Cells 2019, 42, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, Y.H.; Schmidt-Dannert, C.; Lee, P.C. Redesign, reconstruction, and directed extension of the Brevibacterium linens C40 carotenoid pathway in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 5199–5206. [Google Scholar] [CrossRef] [Green Version]
- Song, G.H.; Kim, S.H.; Choi, B.H.; Han, S.J.; Lee, P.C. Heterologous Carotenoid-Biosynthetic Enzymes: Functional Complementation and Effects on Carotenoid Profiles in Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 610–618. [Google Scholar] [CrossRef] [Green Version]
- Cavaleiro, A.M.; Kim, S.H.; Seppala, S.; Nielsen, M.T.; Norholm, M.H. Accurate DNA Assembly and Genome Engineering with Optimized Uracil Excision Cloning. ACS Synth. Biol. 2015, 4, 1042–1046. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Tang, J.; Liu, Y.; Zhu, X.; Zhang, T.; Zhang, X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 2012, 93, 2455–2462. [Google Scholar] [CrossRef]
- Kane, M.A.; Chen, N.; Sparks, S.; Napoli, J.L. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 2005, 15, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, U.; Javarasetty, C.; Murari, S.K. Purification and Fractional Analysis of Methanolic Extract of Wedelia Trilobata Possessing Apoptotic and Anti-Leukemic Activity. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Wahyuni, F.S.; Shaari, K.; Stanslas, J.; Lajis, N.H.; Hamidi, D. Cytotoxic Properties and Complete Nuclear Magnetic Resonance Assignment of Isolated Xanthones from the Root of Garcinia cowa Roxb. Pharmacogn. Mag. 2016, 12, S52–S56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.Y.; Binkley, R.M.; Kim, W.J.; Lee, M.H.; Lee, S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab. Eng. 2018, 49, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Xu, H.; Yu, H. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Goodman, D.B.; Church, G.M.; Kosuri, S. Causes and effects of N-terminal codon bias in bacterial genes. Science 2013, 342, 475–479. [Google Scholar] [CrossRef] [Green Version]
- Jarboe, L.R. YqhD: A broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol. 2011, 89, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Layton, D.S.; Trinh, C.T. Engineering modular ester fermentative pathways in Escherichia coli. Metab. Eng. 2014, 26, 77–88. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef]
- Bienick, M.S.; Young, K.W.; Klesmith, J.R.; Detwiler, E.E.; Tomek, K.J.; Whitehead, T.A. The interrelationship between promoter strength, gene expression, and growth rate. PLoS ONE 2014, 9, e109105. [Google Scholar] [CrossRef]
- Ruiz, A.; Winston, A.; Lim, Y.H.; Gilbert, B.A.; Rando, R.R.; Bok, D. Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem. 1999, 274, 3834–3841. [Google Scholar] [CrossRef] [Green Version]
- Van Breemen, R.B.; Nikolic, D.; Xu, X.; Xiong, Y.; van Lieshout, M.; West, C.E.; Schilling, A.B. Development of a method for quantitation of retinol and retinyl palmitate in human serum using high-performance liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry. J. Chromatogr. A 1998, 794, 245–251. [Google Scholar] [CrossRef]
- Fu, P.P.; Xia, Q.; Yin, J.J.; Cherng, S.H.; Yan, J.; Mei, N.; Chen, T.; Boudreau, M.D.; Howard, P.C.; Wamer, W.G. Photodecomposition of Vitamin A and Photobiological Implications for the Skin. Photochem. Photobiol. 2007, 83, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Jeude, M.; Dittrich, B.; Niederschulte, H.; Anderlei, T.; Knocke, C.; Klee, D.; Buchs, J. Fed-batch mode in shake flasks by slow-release technique. Biotechnol. Bioeng. 2006, 95, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Varani, J.; Warner, R.L.; Gharaee-Kermani, M.; Phan, S.H.; Kang, S.; Chung, J.H.; Wang, Z.Q.; Datta, S.C.; Fisher, G.J.; Voorhees, J.J. Vitamin A antagonizes decreased cell growth and elevated collagen-degrading matrix metalloproteinases and stimulates collagen accumulation in naturally aged human skin. J. Investig. Dermatol. 2000, 114, 480–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamus, J.; Feng, L.; Hawkins, S.; Kalleberg, K.; Lee, J.M. Climbazole boosts activity of retinoids in skin. Int. J. Cosmet. Sci. 2017, 39, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Ahlemeyer, B.; Bauerbach, E.; Plath, M.; Steuber, M.; Heers, C.; Tegtmeier, F.; Krieglstein, J. Retinoic acid reduces apoptosis and oxidative stress by preservation of SOD protein level. Free Radic. Biol. Med. 2001, 30, 1067–1077. [Google Scholar] [CrossRef]






| Strains and Plasmids | Relevant Properties | Source or Reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | F- mcrA Δφ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 | Invitrogen |
| E. coli XL1-Blue | EndA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F′[::Tn10 proAB+ laclq Δ(lacZ)M15] hsdR17 (rK- mK+) | Stratagene |
| E. faecalis Gf2 | Source for idi and ispA | KCTC21002 |
| B. subtilis subsp. spizizenii W23 ATCC 6633 | Source for dxs and dxr | KCTC2189 |
| S. ruber strain M31 | Source for blhSR, brpSR, and bcoxSR | DSM13855 |
| Hep3B cell line | Source for CRBP and LRAT | [25] |
| HS68 | Human foreskin fibroblast cell 68 | ATCC CRL-1635 |
| Recombinant E. coli strains | ||
| XIASR | XL1-Blue ΔglvC::idi ΔyjbI::ispA ΔilvG::Dxs ΔagaVWA::dxr | This study |
| XB | XIASR, pAC-crtEPA-crtBPA-crtIPA-crtYPA | This study |
| XRD1 | XB, pUCMr-blhSR | This study |
| XRD2 | XB, pUCMr-brpSR | This study |
| XRD3 | XB, pUCMr-bcoxSR | This study |
| XRD4 | XB, pUCMr-12blhSR | This study |
| XRD5 | XB, pUCMr-37blhSR | This study |
| XRD6 | XB, pUCMr-46blhSR | This study |
| XRD7 | XB, pUCM-blhSR-CRBPHS-LRATHS | This study |
| XRD8 | XB, pUCM-12blhSR-CRBPHS-LRATHS | This study |
| Plasmids for pathway construction | ||
| pUCM | Cloning vector modified from pUC19; constitutive lac promoter, Amp | [26] |
| pET21a | Source for rop | Novagen |
| pUCMr | Cloning vector modified from pUCM; constitutive lac promoter and rop gene, Amp (low copy plasmid) | This study |
| pUCMr12 | Cloning vector modified from pUCMr; UTR12 sequence (GTTTAAACTGACTGACGCACCAAAAG) | This study |
| pUCMr37 | Cloning vector modified from pUCMr; UTR37 sequence (GTTTAAACAATAAATTACGAGCCAGT) | This study |
| pUCMr46 | Cloning vector modified from pUCMr; UTR46 sequence (GTTTAAACCGAATTGGTGGGGCG) | This study |
| pUCMr-blhSR | Constitutive expressed blh gene from S. ruber | This study |
| pUCMr-brpSR | Constitutive expressed brp gene from S. ruber | This study |
| pUCMr-bcoxSR | Constitutive expressed bcox gene from S. ruber | This study |
| pUCMr-12blhSR | Constitutive expressed blh gene from S. ruber with UTR12 sequence | This study |
| pUCMr-37blhSR | Constitutive expressed blh gene from S. ruber with UTR37 sequence | This study |
| pUCMr-46blhSR | Constitutive expressed blh gene from S. ruber with UTR46 sequence | This study |
| pUCM-LRATHS | Constitutive expressed LRAT gene from Homo Sapiens | This study |
| pUCM-CRBPHS-LRATHS | Constitutive expressed CRBP and LRAT genes from Homo Sapiens | This study |
| pUCM-blhSR-CRBPHS-LRATHS | Constitutive expressed blh, CRBP, and LRAT genes | This study |
| pUCM-12blhSR-CRBPHS-LRATHS | Constitutive expressed 12blh, CRBP, and LRAT genes | This study |
| pAC-EBIY | Constitutive expressed crtE, crtB, crtI, and crtY genes from Pantoea agglomerans | [27] |
| Plasmids for genome engineering | ||
| pUCM-idi | Constitutive expressed idi gene from E. faecalis Gf2 | This study |
| pUCM-ispA | Constitutive expressed ispA gene from E. faecalis Gf2 | This study |
| pUCM-dxs | Constitutive expressed dxs gene from B. subtilis subsp.spizizenii W23 ATCC 6633 | This study |
| pUCM-dxr | Constitutive expressed dxr gene from B. subtilis subsp.spizizenii W23 ATCC 6633 | This study |
| pFRT-idi | Integrative plasmid, glvC_UP-FRT-pPGK-KanR-FRT-idi-glvC_down cassette with ColE1 origin | This study |
| pFRT-ispA | Integrative plasmid, yjbI_UP-FRT-pPGK-KanR-FRT-ispA-yjbI_down cassette with ColE1 origin | This study |
| pFRT-dxs | Integrative plasmid, ilvG_UP-FRT-pPGK-KanR-FRT-Dxs-ilvG_down cassette with ColE1 origin | This study |
| pFRT-dxr | Integrative plasmid, agaVWA_UP-FRT-pPGK-KanR-FRT-Dxr-agaVWA_down cassette with ColE1 origin | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, B.H.; Hwang, H.J.; Lee, J.E.; Oh, S.H.; Hwang, J.S.; Lee, B.Y.; Lee, P.C. Microbial Production of Retinyl Palmitate and Its Application as a Cosmeceutical. Antioxidants 2020, 9, 1130. https://doi.org/10.3390/antiox9111130
Choi BH, Hwang HJ, Lee JE, Oh SH, Hwang JS, Lee BY, Lee PC. Microbial Production of Retinyl Palmitate and Its Application as a Cosmeceutical. Antioxidants. 2020; 9(11):1130. https://doi.org/10.3390/antiox9111130
Chicago/Turabian StyleChoi, Bo Hyun, Hee Jin Hwang, Ji Eun Lee, Soon Hwan Oh, Jae Sung Hwang, Bun Yeoul Lee, and Pyung Cheon Lee. 2020. "Microbial Production of Retinyl Palmitate and Its Application as a Cosmeceutical" Antioxidants 9, no. 11: 1130. https://doi.org/10.3390/antiox9111130