Polyphenols and IUGR Pregnancies: Effects of the Antioxidant Hydroxytyrosol on the Hippocampus Proteome in a Porcine Model
Abstract
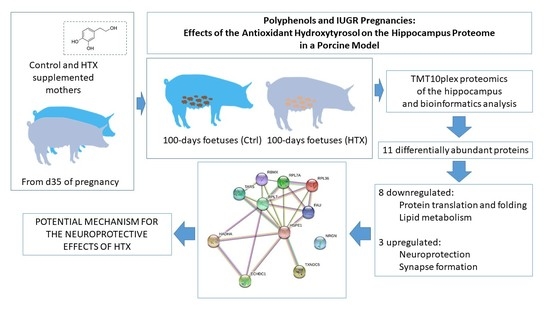
:1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Procedure and Ethics Statement
2.2. Isobaric Mass Tag Labelling with TMT10plex™
2.3. High pH-Reverse Phase (HP-RP) Liquid Chromatography and LC-MS/MS Analysis
2.4. Data Analysis and Quantification
2.5. Gene Ontology and Bioinformatic Analysis
3. Results
3.1. Effects of Maternal Supplementation with HTX on the Foetal Hippocampal Proteome
3.2. Gene Ontology Analysis
3.2.1. Molecular Function
- (A)
- Binding proteins (GO: 0005488; 37.5%), including three proteins that represent several functions: ion binding (GO:0043167; one protein: HSPE1), heterocyclic compounds binding (GO:1901363; two proteins: RPL7A and RPL7), organic compounds binding (GO:0097159; two proteins: RPL7A and RPL7), and other proteins binding (GO:0005515; one protein: HSPE1).
- (B)
- Proteins with catalytic activity (GO:0003824; 37.5%), including three proteins classified as ligases (GO:0016874; one protein: TARS1), lyases (GO:0016829; two proteins: HADHA and ECHDC1), on RNA (GO:0140098; one protein: TARS1), and oxidoreductases (GO:0016491; one protein: HADHA).
- (C)
- Structural Proteins (GO:0005198; 25%), including two proteins corresponding to structural components of the ribosome (GO:0003735; RPL36 and RPL7).
3.2.2. Biological Process
- (A)
- Cellular processes (GO: 0009987; 50%), including eight proteins corresponding mainly to metabolism (GO: 0044237; seven proteins: RPL36, RPL7A, RPL7, TARS1, HADHA, ECHDC1, and RBMX), protein folding (GO:0006457; one protein: HSPE1), and cellular component organization or biogenesis (GO:0071840; two proteins: RPL7A and RPL7).
- (B)
- Metabolic Processes (GO: 0008152; 44%), including seven proteins (RPL36, RPL7A, RPL7, TARS1, HADHA, ECHDC1, and RBMX) that mainly participate in the metabolism of organic compounds (GO: 0071704), cellular metabolism (GO:0044237), and primary metabolism (GO:0044238).
- (C)
- Regulation of biological processes (GO:0065007; 6%) with one protein (RBMX).
3.3. Pathway Analysis with Reactome and KEGG
3.4. Network Analysis with String
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vuguin, P.M. Animal Models for Small for Gestational Age and Fetal Programing of Adult Disease. Horm. Res. 2007, 68, 113–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Bazer, F.W.; Wallace, J.M.; Spencer, T.E. Board-Invited Review: Intrauterine Growth Retardation: Implications for the Animal Sciences. J. Anim. Sci. 2006, 84, 2316–2337. [Google Scholar] [CrossRef] [PubMed]
- Hovi, P.; Andersson, S.; Eriksson, J.G.; Järvenpää, A.-L.; Strang-Karlsson, S.; Mäkitie, O.; Kajantie, E. Glucose Regulation in Young Adults with Very Low Birth Weight. N. Engl. J. Med. 2007, 356, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.G.; Desai, M. Developmental Programming of Offspring Obesity, Adipogenesis, and Appetite. Clin. Obstet. Gynecol. 2013, 56, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Al Wattar, B.H.; Dodds, J.; Placzek, A.; Beresford, L.; Spyreli, E.; Moore, A.; Gonzalez Carreras, F.J.; Austin, F.; Murugesu, N.; Roseboom, T.J.; et al. Mediterranean-Style Diet in Pregnant Women with Metabolic Risk Factors (ESTEEM): A Pragmatic Multicentre Randomised Trial. PLoS Med. 2019, 16, e1002857. [Google Scholar] [CrossRef]
- Martínez-Galiano, J.M.; Olmedo-Requena, R.; Barrios-Rodríguez, R.; Amezcua-Prieto, C.; Bueno-Cavanillas, A.; Salcedo-Bellido, I.; Jimenez-Moleon, J.J.; Delgado-Rodríguez, M. Effect of Adherence to a Mediterranean Diet and Olive Oil Intake during Pregnancy on Risk of Small for Gestational Age Infants. Nutrients 2018, 10, 1234. [Google Scholar] [CrossRef] [Green Version]
- Robles-Almazan, M.; Pulido-Moran, M.; Moreno-Fernandez, J.; Ramirez-Tortosa, C.; Rodriguez-Garcia, C.; Quiles, J.L.; Ramirez-Tortosa, M. Hydroxytyrosol: Bioavailability, Toxicity, and Clinical Applications. Food Res. Int. 2018, 105, 654–667. [Google Scholar] [CrossRef]
- Bertelli, M.; Kiani, A.K.; Paolacci, S.; Manara, E.; Kurti, D.; Dhuli, K.; Bushati, V.; Miertus, J.; Pangallo, D.; Baglivo, M.; et al. Hydroxytyrosol: A Natural Compound with Promising Pharmacological Activities. J. Biotechnol. 2020, 309, 29–33. [Google Scholar] [CrossRef]
- Serreli, G.; Deiana, M. Biological Relevance of Extra Virgin Olive Oil Polyphenols Metabolites. Antioxidants 2018, 7, 170. [Google Scholar] [CrossRef] [Green Version]
- Domínguez-Perles, R.; Auñón, D.; Ferreres, F.; Gil-Izquierdo, A. Physiological Linkage of Gender, Bioavailable Hydroxytyrosol Derivatives, and Their Metabolites with Systemic Catecholamine Metabolism. Food Funct. 2017, 8, 4570. [Google Scholar] [CrossRef]
- Zheng, A.; Li, H.; Cao, K.; Xu, J.; Zou, X.; Li, Y.; Chen, C.; Liu, J.; Feng, Z. Maternal Hydroxytyrosol Administration Improves Neurogenesis and Cognitive Function in Prenatally Stressed Offspring. J. Nutr. Biochem. 2015, 26, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Leri, M.; Scuto, M.; Ontario, M.L.; Calabrese, V.; Calabrese, E.J.; Bucciantini, M.; Stefani, M. Healthy Effects of Plant Polyphenols: Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varghese, N.; Werner, S.; Grimm, A.; Eckert, A. Dietary Mitophagy Enhancer: A Strategy for Healthy Brain Aging? Antioxidants 2020, 9, 932. [Google Scholar] [CrossRef] [PubMed]
- De Pablos, R.M.; Espinosa-Oliva, A.M.; Hornedo-Ortega, R.; Cano, M.; Arguelles, S. Hydroxytyrosol Protects from Aging Process via AMPK and Autophagy; A Review of Its Effects on Cancer, Metabolic Syndrome, Osteoporosis, Immune-Mediated and Neurodegenerative Diseases. Pharmacol. Res. 2019, 143, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Robbins, M.E.; Revhaug, C.; Saugstad, O.D. Oxygen Radical Disease in the Newborn, Revisited: Oxidative Stress and Disease in the Newborn Period. Free Radic. Biol. Med. 2019, 142, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Rashid, C.S.; Bansal, A.; Simmons, R.A. Oxidative Stress, Intrauterine Growth Restriction, and Developmental Programming of Type 2 Diabetes. Physiology 2018, 33, 348–359. [Google Scholar] [CrossRef] [Green Version]
- Aljunaidy, M.M.; Morton, J.S.; Cooke, C.L.M.; Davidge, S.T. Prenatal Hypoxia and Placental Oxidative Stress: Linkages to Developmental Origins of Cardiovascular Disease. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R395–R399. [Google Scholar] [CrossRef]
- Richter, H.G.; Camm, E.J.; Modi, B.N.; Naeem, F.; Cross, C.M.; Cindrova-Davies, T.; Spasic-Boskovic, O.; Dunster, C.; Mudway, I.S.; Kelly, F.J.; et al. Ascorbate Prevents Placental Oxidative Stress and Enhances Birth Weight in Hypoxic Pregnancy in Rats. J. Physiol. 2012, 590, 1377–1387. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, W.; Liu, J. Neurodevelopment in Children with Intrauterine Growth Restriction: Adverse Effects and Interventions. J. Matern. Neonatal Med. 2016, 29, 660–668. [Google Scholar] [CrossRef]
- Yzydorczyk, C.; Armengaud, J.B.; Peyter, A.C.; Chehade, H.; Cachat, F.; Juvet, C.; Siddeek, B.; Simoncini, S.; Sabatier, F.; Dignat-George, F.; et al. Endothelial Dysfunction in Individuals Born after Fetal Growth Restriction: Cardiovascular and Renal Consequences and Preventive Approaches. J. Dev. Orig. Health Dis. 2017, 8, 448–464. [Google Scholar] [CrossRef]
- Gonzalez-Bulnes, A.; Ovilo, C.; Lopez-Bote, C.J.; Astiz, S.; Ayuso, M.; Perez-Solana, M.; Sanchez-Sanchez, R.; Torres-Rovira, L. Gender-Specific Early Postnatal Catch-up Growth after Intrauterine Growth Retardation by Food Restriction in Swine with Obesity/Leptin Resistance. Reproduction 2012, 144, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Bulnes, A.; Astiz, S.; Parraguez, V.H.; Garcia-Contreras, C.; Vazquez-Gomez, M. Empowering Translational Research in Fetal Growth Restriction: Sheep and Swine Animal Models. Curr. Pharm. Biotechnol. 2016, 17, 848–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Gómez, M.; Valent, D.; García-Contreras, C.; Arroyo, L.; Óvilo, C.; Isabel, B.; Bassols, A.; González-Bulnes, A. Sex and Intrauterine Growth Restriction Modify Brain Neurotransmitters Profile of Newborn Piglets. Int. J. Dev. Neurosci. 2016, 55, 9–14. [Google Scholar] [CrossRef] [PubMed]
- García-Contreras, C.; Valent, D.; Vázquez-Gómez, M.; Arroyo, L.; Isabel, B.; Astiz, S.; Bassols, A.; Gonzalez-Bulnes, A. Fetal Growth-Retardation and Brain-Sparing by Malnutrition Are Associated to Changes in Neurotransmitters Profile. Int. J. Dev. Neurosci. 2017, 57, 72–76. [Google Scholar] [CrossRef]
- Valent, D.; Yeste, N.; Hernández-Castellano, L.E.; Arroyo, L.; Wu, W.; García-Contreras, C.; Vázquez-Gómez, M.; González-Bulnes, A.; Bendixen, E.; Bassols, A. SWATH-MS Quantitative Proteomic Investigation of Intrauterine Growth Restriction in a Porcine Model Reveals Sex Differences in Hippocampus Development. J. Proteomics 2019, 204, 103391. [Google Scholar] [CrossRef]
- Wixey, J.A.; Lee, K.M.; Miller, S.M.; Goasdoue, K.; Colditz, P.B.; Tracey Bjorkman, S.; Chand, K.K. Neuropathology in Intrauterine Growth Restricted Newborn Piglets Is Associated with Glial Activation and Proinflammatory Status in the Brain 11 Medical and Health Sciences 1109 Neurosciences. J. Neuroinflamm. 2019, 16, 5. [Google Scholar] [CrossRef]
- Miller, S.S.L.; Huppi, P.S.P.; Mallard, C. The Consequences of Fetal Growth Restriction on Brain Structure and Neurodevelopmental Outcome. J. Physiol. 2016, 594, 807–823. [Google Scholar] [CrossRef] [Green Version]
- Cumberland, A.L.; Palliser, H.K.; Rani, P.; Walker, D.W.; Hirst, J.J. Effects of Combined IUGR and Prenatal Stress on the Development of the Hippocampus in a Fetal Guinea Pig Model. J. Dev. Orig. Health Dis. 2017, 8, 584–596. [Google Scholar] [CrossRef]
- Bauer, R.; Walter, B.; Vorwieger, G.; Bergmann, R.; Füchtner, F.; Brust, P. Intrauterine Growth Restriction Induces Up-Regulation of Cerebral Aromatic Amino Acid Decarboxylase Activity in Newborn Piglets: [18F]Fluorodopa Positron Emission Tomographic Study. Pediatr. Res. 2001, 49, 474–480. [Google Scholar] [CrossRef] [Green Version]
- Bauer, R.; Walter, B.; Brust, P.; Füchtner, F.; Zwiener, U. Impact of Asymmetric Intrauterine Growth Restriction on Organ Function in Newborn Piglets. Eur. J. Obstet. Gynecol. Reprod. Biol. 2003, 110, S40–S49. [Google Scholar] [CrossRef]
- Hernández-Andrade, E.; Cortés-Camberos, A.J.; Díaz, N.F.; Flores-Herrera, H.; García-López, G.; González-Jiménez, M.; Santamaría, A.; Molina-Hernández, A. Altered Levels of Brain Neurotransmitter from New Born Rabbits with Intrauterine Restriction. Neurosci. Lett. 2015, 584, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, Z.; Totoki, K.; Schoch, H.; Whitaker, K.W.; Hill-Smith, T.; Lucki, I.; Reyes, T.M. Early Life Protein Restriction Alters Dopamine Circuitry. Neuroscience 2010, 168, 359–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgane, P.J.; Mokler, D.J.; Galler, J.R. Effects of Prenatal Protein Malnutrition on the Hippocampal Formation. Neurosci. Biobehav. Rev. 2002, 26, 471–483. [Google Scholar] [CrossRef]
- Vazquez-Gomez, M.; Garcia-Contreras, C.; Torres-Rovira, L.; Pesantez, J.L.; Gonzalez-Añover, P.; Gomez-Fidalgo, E.; Sanchez-Sanchez, R.; Ovilo, C.; Isabel, B.; Astiz, S.; et al. Polyphenols and IUGR Pregnancies: Maternal Hydroxytyrosol Supplementation Improves Prenatal and Early-Postnatal Growth and Metabolism of the Offspring. PLoS ONE 2017, 12, e0177593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez-Gomez, M.; Heras-Molina, A.; Garcia-Contreras, C.; Pesantez-Pacheco, J.L.; Torres-Rovira, L.; Martinez-Fernandez, B.; Gonzalez, J.; Encinas, T.; Astiz, S.; Ovilo, C.; et al. Polyphenols and IUGR Pregnancies: Effects of Maternal Hydroxytyrosol Supplementation on Postnatal Growth, Metabolism and Body Composition of the Offspring. Antioxidants 2019, 8, 535. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Contreras, C.; Vazquez-Gomez, M.; Barbero, A.; Pesantez, J.L.; Zinellu, A.; Berlinguer, F.; Gonzalez-Añover, P.; Gonzalez, J.; Encinas, T.; Torres-Rovira, L.; et al. Polyphenols and Iugr Pregnancies: Effects of Maternal Hydroxytyrosol Supplementation on Placental Gene Expression and Fetal Antioxidant Status, Dna-Methylation and Phenotype. Int. J. Mol. Sci. 2019, 20, 1187. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Contreras, C.; Vazquez-Gomez, M.; Pardo, Z.; Heras-Molina, A.; Pesantez, J.L.; Encinas, T.; Torres-Rovira, L.; Astiz, S.; Nieto, R.; Ovilo, C.; et al. Polyphenols and IUGR Pregnancies: Effects of Maternal Hydroxytyrosol Supplementation on Hepatic Fat Accretion and Energy and Fatty Acids Profile of Fetal Tissues. Nutrients 2019, 11, 1534. [Google Scholar] [CrossRef] [Green Version]
- Yeste, N.; Valent, D.; Arroyo, L.; Vázquez-Gómez, M.; García-Contreras, C.; Pumarola, M.; González-Bulnes, A.; Bassols, A. Polyphenols and IUGR Pregnancies: Effects of the Antioxidant Hydroxytyrosol on Brain Neurochemistry and Development in a Porcine Model. Antioxidants 2021, 10, 884–905. [Google Scholar] [CrossRef]
- Yeste, N.; Gómez, N.; Vázquez-Gómez, M.; García-Contreras, C.; Pumarola, M.; González-Bulnes, A.; Bassols, A. Polyphenols and IUGR Pregnancies: Intrauterine Growth Restriction and Hydroxytyrosol Affect the Development and Neurotransmitter Profile of the Hippocampus in a Pig Model. Antioxidants 2021, 10, 1505. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER Version 11: Expanded Annotation Data from Gene Ontology and Reactome Pathways, and Data Analysis Tool Enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Sci. 2019, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, L.; Valent, D.; Carreras, R.; Pato, R.; Sabrià, J.; Velarde, A.; Bassols, A. Neurobiology of Environmental Enrichment in Pigs: Hanges in Monoaminergic Neurotransmitters in Several Brain Areas and in the Hippocampal Proteome. J. Proteomics 2020, 229, 103943. [Google Scholar] [CrossRef] [PubMed]
- Linster, C.L.; Noël, G.; Stroobant, V.; Vertommen, D.; Vincent, M.F.; Bommer, G.T.; Veiga-da-Cunha, M.; Van Schaftingen, E. Ethylmalonyl-CoA Decarboxylase, a New Enzyme Involved in Metabolite Proofreading. J. Biol. Chem. 2011, 286, 42992–43003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewulf, J.P.; Paquay, S.; Marbaix, E.; Achouri, Y.; van Schaftingen, E.; Bommer, G.T. ECHDC1 Knockout Mice Accumulate Ethyl-Branched Lipids and Excrete Abnormal Intermediates of Branched-Chain Fatty Acid Metabolism. J. Biol. Chem. 2021, 297, 101083. [Google Scholar] [CrossRef]
- Horna-Terrón, E.; Pradilla-Dieste, A.; Sánchez-de-Diego, C.; Osada, J. TXNDC5, a Newly Discovered Disulfide Isomerase with a Key Role in Cell Physiology and Pathology. Int. J. Mol. Sci. 2014, 15, 23501–23518. [Google Scholar] [CrossRef] [Green Version]
- Díez-Guerra, F.J. Neurogranin, a Link between Calcium/Calmodulin and Protein Kinase C Signaling in Synaptic Plasticity. IUBMB Life 2010, 62, 597–606. [Google Scholar] [CrossRef]
- Miyakawa, T.; Yared, E.; Pak, J.H.; Huang, F.L.; Huang, K.P.; Crawley, J.N. Neurogranin Null Mutant Mice Display Performance Deficits on Spatial Learning Tasks with Anxiety Related Components. Hippocampus 2001, 11, 763–775. [Google Scholar] [CrossRef]
- Bie, A.S.; Fernandez-Guerra, P.; Birkler, R.I.D.; Nisemblat, S.; Pelnena, D.; Lu, X.; Deignan, J.L.; Lee, H.; Dorrani, N.; Corydon, T.J.; et al. Effects of a Mutation in the HSPE1 Gene Encoding the Mitochondrial Co-Chaperonin HSP10 and Its Potential Association with a Neurological and Developmental Disorder. Front. Mol. Biosci. 2016, 3, 65. [Google Scholar] [CrossRef]
- Rajendran, V.; Kalita, P.; Shukla, H.; Kumar, A.; Tripathi, T. Aminoacyl-TRNA Synthetases: Structure, Function, and Drug Discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002, 12, 1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanhoush, R.; Beenders, B.; Perrin, C.; Moreau, J.; Bellini, M.; Penrad-Mobayed, M. Novel Domains in the HnRNP G/RBMX Protein with Distinct Roles in RNA Binding and Targeting Nascent Transcripts. Nucleus 2010, 1, 109–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of Energy Metabolism by Long-Chain Fatty Acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Schönfeld, P.; Reiser, G. Why Does Brain Metabolism Not Favor Burning of Fatty Acids to Provide Energy? Reflections on Disadvantages of the Use of Free Fatty Acids as Fuel for Brain. J. Cereb. Blood Flow Metab. 2013, 33, 1493–1499. [Google Scholar] [CrossRef] [Green Version]


| Reporter | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | 131 | |
| Experiment 1 | CF.1 | CM.5 | CF.2 | TM.1 | TF.5 | CF.3 | CM.4 | TF.4 | TM.2 | TF.3 |
| Experiment 2 | CM.3 | TF.2 | TM.3 | CF.4 | CM.2 | TM.4 | TF.1 | CM.1 | CF.5 | TM.5 |
| Access Uniprot | Gen | Identification | FC | p-Value |
|---|---|---|---|---|
| F1SMZ6 | HSPE1 | 10 kDa heat shock protein, mitochondrial | 0.06 | 0.048 |
| F1SP18 | TARS1 | Threonyl-tRNA synthetase | 0.50 | 0.019 |
| P62863 | FAU | 40S ribosomal protein S30 | 0.52 | 0.002 |
| Q29375 | RPL7A | 60S ribosomal protein L7a | 0.56 | 0.007 |
| Q29554 | HADHA | Trifunctional enzyme subunit alpha, mitochondrial | 0.57 | <0.001 |
| P83884 | RPL36 | 60S ribosomal protein L36 | 0.60 | 0.012 |
| A5GFQ0 | RPL7 | 60S ribosomal protein L7 | 0.60 | 0.004 |
| F1RQ90 | RBMX | RNA-binding motif protein, X chromosome isoform 1 | 0.65 | <0.001 |
| F1S2 × 3 | ECHDC1 | Ethylmalonyl-CoA decarboxylase 1 | 1.53 | 0.049 |
| A0A287ARZ1 | TXNDC5 | Thioredoxin domain-containing protein 5 | 1.73 | 0.020 |
| A0A287A6U0 | NRGN | Neurogranin | 2.22 | 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeste, N.; Pérez-Valle, J.; Vázquez-Gómez, M.; García-Contreras, C.; González-Bulnes, A.; Bassols, A. Polyphenols and IUGR Pregnancies: Effects of the Antioxidant Hydroxytyrosol on the Hippocampus Proteome in a Porcine Model. Antioxidants 2022, 11, 1135. https://doi.org/10.3390/antiox11061135
Yeste N, Pérez-Valle J, Vázquez-Gómez M, García-Contreras C, González-Bulnes A, Bassols A. Polyphenols and IUGR Pregnancies: Effects of the Antioxidant Hydroxytyrosol on the Hippocampus Proteome in a Porcine Model. Antioxidants. 2022; 11(6):1135. https://doi.org/10.3390/antiox11061135
Chicago/Turabian StyleYeste, Natalia, Jorge Pérez-Valle, Marta Vázquez-Gómez, Consolación García-Contreras, Antonio González-Bulnes, and Anna Bassols. 2022. "Polyphenols and IUGR Pregnancies: Effects of the Antioxidant Hydroxytyrosol on the Hippocampus Proteome in a Porcine Model" Antioxidants 11, no. 6: 1135. https://doi.org/10.3390/antiox11061135