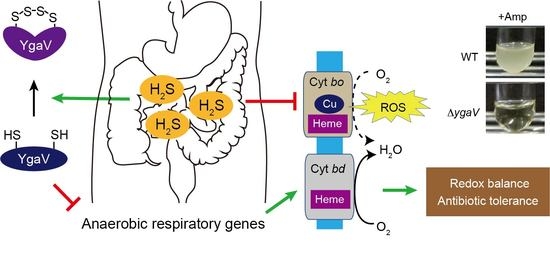
The Sulfide-Responsive SqrR/BigR Homologous Regulator YgaV of Escherichia coli Controls Expression of Anaerobic Respiratory Genes and Antibiotic Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. RNA Sequencing
2.3. Quantitative Real-Time PCR
2.4. Expression and Purification of YgaV
2.5. Reconstitution of Heme-Bound YgaV
2.6. Electrophoretic Mobility Shift Assay
2.7. DNase Footprint Assay
2.8. In Vitro AMS Modification
2.9. Ionization-Associated Cleavage of Oxidized Polysulfur (iCOPS) Method
2.10. Primer Extension Analysis
2.11. Construction of lacZ Fusion and β-Galactosidase Assay
2.12. Measurement of the H2O2 Content
2.13. Antibiotic Susceptibility Test
2.14. Statistics and Reproducibility
2.15. Accession Numbers
3. Results
3.1. Identification of Genes Controlled by YgaV
3.2. Biochemical Property of YgaV
3.3. Thiol-Modification of YgaV and Its Impact on DNA Binding
3.4. Detection of Oxidized Polysulfur Structure of YgaV by iCOPS Method
3.5. YgaV Regulates Transcription in a Sulfide-Dependent Manner
3.6. Phenotype of the YgaV Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sikalidis, A.K. Amino acids and immune response: A role for cysteine, glutamine, phenylalanine, tryptophan and arginine in t-cell function and cancer? Pathol. Oncol. Res. 2015, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Sikalidis, A.K.; Maykish, A. The gut microbiome and type 2 diabetes mellitus: Discussing a complex relationship. Biomedicines 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1997, 1320, 217–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunsalus, R.; Park, S.-J. Aerobic-anaerobic gene regulation in Escherichia coli: Control by the ArcAB and Fnr regulons. Res. Microbiol. 1994, 145, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Toledo, J.C., Jr.; Augusto, O. Connecting the chemical and biological properties of nitric oxide. Chem. Res. Toxicol. 2012, 25, 975–989. [Google Scholar] [CrossRef]
- Walsh, B.J.C.; Giedroc, D.P. H2S and reactive sulfur signaling at the host-bacterial pathogen interface. J. Biol. Chem. 2020, 295, 13150–13168. [Google Scholar] [CrossRef]
- Kimura, H. Production and physiological effects of hydrogen sulfide. Antioxid. Redox Signal. 2014, 20, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Fujii, S.; Matsunaga, T.; Nishimura, A.; Ono, K.; Ida, T.; Ahmed, K.A.; Okamoto, T.; Tsutsuki, H.; Sawa, T.; et al. Reactive persulfides from Salmonella typhimurium downregulate autophagy-mediated innate immunity in macrophages by inhibiting electrophilic signaling. Cell Chem. Biol. 2018, 25, 1403–1413.e4. [Google Scholar] [CrossRef]
- Nishida, M.; Sawa, T.; Kitajima, N.; Ono, K.; Inoue, H.; Ihara, H.; Motohashi, H.; Yamamoto, M.; Suematsu, M.; Kurose, H.; et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012, 8, 714–724. [Google Scholar] [CrossRef]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and s-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [Green Version]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P.; Kantor, R.S.; Lavy, A.; Warren, L.A.; Rappé, M.S.; Pester, M.; Loy, A.; Thomas, B.C.; et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Gibson, G.R.; Cummings, J.H. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992, 72, 57–64. [Google Scholar] [CrossRef]
- Wang, J.; Guo, X.; Li, H.; Qi, H.; Qian, J.; Yan, S.; Shi, J.; Niu, W. Hydrogen sulfide from cysteine desulfurase, not 3-mercaptopyruvate sulfurtransferase, contributes to sustaining cell growth and bioenergetics in E. coli under anaerobic conditions. Front. Microbiol. 2019, 10, 2357. [Google Scholar] [CrossRef]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef]
- Mironov, A.; Seregina, T.; Nagornykh, M.; Luhachack, L.G.; Korolkova, N.; Lopes, E.L.; Kotova, V.; Zavilgelsky, G.; Shakulov, R.; Shatalin, K.; et al. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 6022–6027. [Google Scholar] [CrossRef] [Green Version]
- Shukla, P.; Khodade, V.S.; SharathChandra, M.; Chauhan, P.; Mishra, S.; Siddaramappa, S.; Pradeep, B.E.; Singh, A.; Chakrapani, H. “On Demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem. Sci. 2017, 8, 4967–4972. [Google Scholar] [CrossRef] [Green Version]
- Luhachack, L.; Nudler, E. Bacterial gasotransmitters: An innate defense against antibiotics. Curr. Opin. Microbiol. 2014, 21, 13–17. [Google Scholar] [CrossRef]
- Korshunov, S.; Imlay, K.; Imlay, J.A. The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol. Microbiol. 2016, 101, 62–77. [Google Scholar] [CrossRef] [Green Version]
- Lobritz, M.A.; Belenky, P.; Porter, C.B.M.; Gutierrez, A.; Yang, J.H.; Schwarz, E.G.; Dwyer, D.J.; Khalil, A.S.; Collins, J.J. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef]
- Beebout, C.J.; Sominsky, L.A.; Eberly, A.R.; Van Horn, G.T.; Hadjifrangiskou, M. Cytochrome bd promotes Escherichia coli biofilm antibiotic tolerance by regulating accumulation of noxious chemicals. NPJ Biofilms Microbiomes 2021, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Kitamura, Y.; Zhang, T.; Tsutsuki, H.; Rahman, A.; Ihara, T.; Akaike, T.; Sawa, T. Cysteine hydropersulfide inactivates β-lactam antibiotics with formation of ring-opened carbothioic s-acids in bacteria. ACS Chem. Biol. 2021, 16, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Saini, V.; Chinta, K.C.; Reddy, V.P.; Glasgow, J.N.; Stein, A.; Lamprecht, D.A.; Rahman, M.A.; Mackenzie, J.S.; Truebody, B.E.; Adamson, J.H.; et al. Hydrogen sulfide stimulates mycobacterium tuberculosis respiration, growth and pathogenesis. Nat. Commun. 2020, 11, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Shen, J.; Fang, M.; Zhang, Y.; Hori, K.; Trinidad, J.C.; Bauer, C.E.; Giedroc, D.P.; Masuda, S. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 2355–2360. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, R.L.; Benedetti, C.E. BigR, a Transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth. J. Bacteriol. 2007, 189, 6185–6194. [Google Scholar] [CrossRef] [Green Version]
- De Lira, N.P.V.; Pauletti, B.A.; Marques, A.C.; Perez, C.A.; Caserta, R.; de Souza, A.A.; Vercesi, A.E.; Paes Leme, A.F.; Benedetti, C.E. BigR is a sulfide sensor that regulates a sulfur transferase/dioxygenase required for aerobic respiration of plant bacteria under sulfide stress. Sci. Rep. 2018, 8, 3508. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, B.G.; Barbosa, R.L.; Soprano, A.S.; Campos, B.M.; de Souza, T.A.; Tonoli, C.C.; Leme, A.; Murakami, M.T.; Benedetti, C.E. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 2011, 286, 26148–26157. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Masuda, S. Persulphide-responsive transcriptional regulation and metabolism in bacteria. J. Biochem. 2020, 167, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, D.A.; Walsh, B.J.C.; Zhang, Y.; Dietrich, C.; Gonzalez-Gutierrez, G.; Giedroc, D.P. Structural basis for persulfide-sensing specificity in a transcriptional regulator. Nat. Chem. Biol. 2020, 17, 65–70. [Google Scholar] [CrossRef]
- Walsh, B.J.C.; Wang, J.; Edmonds, K.A.; Palmer, L.D.; Zhang, Y.; Trinidad, J.C.; Skaar, E.P.; Giedroc, D.P. The response of Acinetobacter baumannii to hydrogen sulfide reveals two independent persulfide-sensing systems and a connection to biofilm regulation. MBio 2020, 11, e01254-20. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli k-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buurman, E.T.; Kim, K.T.; Epstein, W. Genetic evidence for two sequentially occupied K+ binding sites in the Kdp transport ATPase. J. Biol. Chem. 1995, 270, 6678–6685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An r package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef] [Green Version]
- Grenier, F.; Matteau, D.; Baby, V.; Rodrigue, S. Complete genome sequence of Escherichia coli BW25113. Genome Announc. 2014, 2, 1038–1052. [Google Scholar] [CrossRef] [Green Version]
- Glasner, J.D.; Liss, P.; Plunkett, G., III; Darling, A.; Prasad, T.; Rusch, M.; Byrnes, A.; Gilson, M.; Biehl, B.; Blattner, F.R.; et al. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 2003, 31, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Motohashi, K. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 2015, 15, 47. [Google Scholar] [CrossRef] [Green Version]
- Masuda, S.; Tomida, Y.; Ohta, H.; Takamiya, K. The critical role of a hydrogen bond between Gln63 and Trp104 in the blue-light sensing BLUF domain that controls AppA activity. J. Mol. Biol. 2007, 368, 1223–1230. [Google Scholar] [CrossRef]
- Lee, J.K.; Kaplan, S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides: Analysis of the cis-acting downstream regulatory sequence. J. Biol. Chem. 1995, 270, 20453–20458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J. Experiments in Molecular Genetcis. In Assay of β-Galactosidase; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972; ISBN 9780879691066. [Google Scholar]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Gueuné, H.; Durand, M.J.; Thouand, G.; DuBow, M.S. The ygaVP genes of Escherichia coli form a tributyltin-inducible operon. Appl. Environ. Microbiol. 2008, 74, 1954–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmon, K.; Hung, S.P.; Mekjian, K.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 2003, 278, 29837–29855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmon, K.A.; Hung, S.-P.; Steffen, N.R.; Krupp, R.; Baldi, P.; Hatfield, G.W.; Gunsalus, R.P. Global gene expression profiling in Escherichia coli K12: Effects of oxygen availability and ArcA. J. Biol. Chem. 2005, 280, 15084–15096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Hayashi, Y.; Arai, M.; McGlynn, S.E.; Masuda, T.; Masuda, S. Repressor activity of SqrR, a master regulator of persulfide-responsive genes, is regulated by heme coordination. Plant Cell Physiol. 2021, 62, 100–110. [Google Scholar] [CrossRef]
- Tartaglia, L.A.; Storz, G.; Ames, B.N. Identification and molecular analysis of OxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 1989, 210, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Akaike, T.; Ida, T.; Wei, F.-Y.; Nishida, M.; Kumagai, Y.; Alam, M.M.; Ihara, H.; Sawa, T.; Matsunaga, T.; Kasamatsu, S.; et al. Cysteinyl-TRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017, 8, 1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasamatsu, S.; Ida, T.; Koga, T.; Asada, K.; Motohashi, H.; Ihara, H.; Akaike, T. High-precision sulfur metabolomics innovated by a new specific probe for trapping reactive sulfur species. Antioxid. Redox Signal. 2021, 34, 1407–1419. [Google Scholar] [CrossRef]
- Hamid, H.A.; Tanaka, A.; Ida, T.; Nishimura, A.; Matsunaga, T.; Fujii, S.; Morita, M.; Sawa, T.; Fukuto, J.M.; Nagy, P.; et al. Polysulfide stabilization by tyrosine and hydroxyphenyl-containing derivatives that is important for a reactive sulfur metabolomics analysis. Redox Biol. 2019, 21, 101096. [Google Scholar] [CrossRef]
- Bogdándi, V.; Ida, T.; Sutton, T.R.; Bianco, C.; Ditrói, T.; Koster, G.; Henthorn, H.A.; Minnion, M.; Toscano, J.P.; van der Vliet, A.; et al. Speciation of reactive sulfur species and their reactions with alkylating agents: Do we have any clue about what is present inside the cell? Br. J. Pharmacol. 2019, 176, 646–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Imlay, J.A. Cell death from antibiotics without the involvement of reactive oxygen species. Science 2013, 339, 1210–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, C.P.; Albrecht, J.; Gunsalus, R.P. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 1996, 178, 1094–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keren, I.; Wu, Y.; Inocencio, J.; Mulcahy, L.R.; Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013, 339, 1213–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Acker, H.; Gielis, J.; Acke, M.; Cools, F.; Cos, P.; Coenye, T. The role of reactive oxygen species in antibiotic-induced cell death in Burkholderia cepacia complex bacteria. PLoS ONE 2016, 11, e0159837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathai, J.C.; Missner, A.; Kügler, P.; Saparov, S.M.; Zeidel, M.L.; Lee, J.K.; Pohl, P. No facilitator required for membrane transport of hydrogen sulfide. Proc. Natl. Acad. Sci. USA 2009, 106, 16633–16638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatalin, K.; Nuthanakanti, A.; Kaushik, A.; Shishov, D.; Peselis, A.; Shamovsky, I.; Pani, B.; Lechpammer, M.; Vasilyev, N.; Shatalina, E.; et al. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science 2021, 372, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F. Characterization of two novel proteins containing the rhodanese homology domain: YgaP and YbbB of Escherichia coli. Ph.D. Thesis, Viginia Polytechnic Institute and State University, Blackburg, VA, USA, 2003. [Google Scholar]
- Mishanina, T.V.; Libiad, M.; Banerjee, R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 2015, 11, 457–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Lü, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017, 11, 2754–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawa, T.; Ono, K.; Tsutsuki, H.; Zhang, T.; Ida, T.; Nishida, M.; Akaike, T. Reactive cysteine persulphides: Occurrence, biosynthesis, antioxidant activity, methodologies, and bacterial persulphide signalling. Adv. Microb. Physiol. 2018, 72, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Fukuto, J.M.; Ignarro, L.J.; Nagy, P.; Wink, D.A.; Kevil, C.G.; Feelisch, M.; Cortese-Krott, M.M.; Bianco, C.L.; Kumagai, Y.; Hobbs, A.J.; et al. Biological hydropersulfides and related polysulfides—A new concept and perspective in redox biology. FEBS Lett. 2018, 592, 2140–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, N.; Yan, Z.; Fan, K.; Li, H.; Zhao, R.; Xia, Y.; Xun, L.; Liu, H. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol. 2019, 26, 101293. [Google Scholar] [CrossRef] [PubMed]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasubramanian, R.; Hori, K.; Shimizu, T.; Kasamatsu, S.; Okamura, K.; Tanaka, K.; Ihara, H.; Masuda, S. The Sulfide-Responsive SqrR/BigR Homologous Regulator YgaV of Escherichia coli Controls Expression of Anaerobic Respiratory Genes and Antibiotic Tolerance. Antioxidants 2022, 11, 2359. https://doi.org/10.3390/antiox11122359
Balasubramanian R, Hori K, Shimizu T, Kasamatsu S, Okamura K, Tanaka K, Ihara H, Masuda S. The Sulfide-Responsive SqrR/BigR Homologous Regulator YgaV of Escherichia coli Controls Expression of Anaerobic Respiratory Genes and Antibiotic Tolerance. Antioxidants. 2022; 11(12):2359. https://doi.org/10.3390/antiox11122359
Chicago/Turabian StyleBalasubramanian, Rajalakshmi, Koichi Hori, Takayuki Shimizu, Shingo Kasamatsu, Kae Okamura, Kan Tanaka, Hideshi Ihara, and Shinji Masuda. 2022. "The Sulfide-Responsive SqrR/BigR Homologous Regulator YgaV of Escherichia coli Controls Expression of Anaerobic Respiratory Genes and Antibiotic Tolerance" Antioxidants 11, no. 12: 2359. https://doi.org/10.3390/antiox11122359