Distribution and Functions of Monodehydroascorbate Reductases in Plants: Comprehensive Reverse Genetic Analysis of Arabidopsis thaliana Enzymes
Abstract
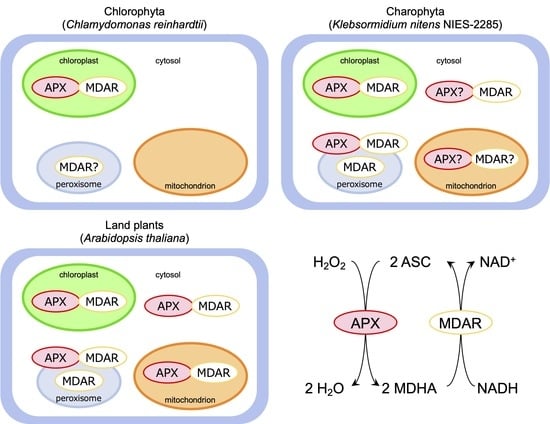
:1. Introduction
2. Materials and Methods
2.1. Mining MDAR Sequences and Phylogenetic Tree Construction
2.2. Plant Materials and Growth Conditions
2.3. Quantitative and Semi-Quantitative Reverse Transcription-PCR Experiments
2.4. Enzyme Assays
2.5. Ascorbate Measurement
2.6. Data Analyses
3. Results
3.1. Mining and Classification of MDAR Isoforms from Green Algae and Land Plants
3.1.1. Mining of MDAR Sequences
3.1.2. Classification of MDAR Isoforms
3.1.3. Class I Chloroplastic/Mitochondrial Enzymes
3.1.4. Class II Peroxisomal Membrane-Associated Enzymes
3.1.5. Class III Cytosolic/Peroxisomal Enzymes
3.1.6. Atypical MDAR Isoforms
3.2. Characterization of Arabidopsis thaliana Mutants Lacking AthMDARs
3.3. Impacts of Class I AthMDAR5 on Ascorbate Pool Size Regulation under Light Stress Conditions
3.4. Complete Loss-of-Function of Both AthMDAR1 and 4 May Cause Lethality
3.5. Neither AthMDAR1 nor AthAPX3 Is Required for Autotrophic Seedling Growth
3.6. Negligible Impacts of AthMDAR1 (Class III) and AthMDAR4 (Class II) on Ascorbate Pool Size Regulation under Light Stress Conditions
3.7. Combined Impacts of Cytosolic AthMDARs and AthDHARs on Ascorbate Pool Size Regulation and Stress Resistance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef]
- Terai, Y.; Ueno, H.; Ogawa, T.; Sawa, Y.; Miyagi, A.; Kawai-Yamada, M.; Ishikawa, T.; Maruta, T. Dehydroascorbate reductases and glutathione set a threshold for high-light-induced ascorbate accumulation. Plant Physiol. 2020, 183, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, B.; Smirnoff, N.; Cobbett, C.S.; Golz, J.F. Ascorbate-deficient vtc2 mutants in Arabidopsis do not exhibit decreased growth. Front. Plant Sci. 2016, 7, 1025. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Shigeoka, S.; Maruta, T. Cellular redox regulation, signaling, and stress response in plants. Biosci. Biotechnol. Biochem. 2014, 78, 1457–1470. [Google Scholar] [CrossRef]
- Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Diversity and evolution of ascorbate peroxidase functions in chloroplasts: More than just a classical antioxidant enzyme? Plant Cell Physiol. 2016, 57, 1377–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignolet-Spruyt, L.; Xu, E.; Idanheimo, N.; Hoeberichts, F.A.; Muhlenbock, P.; Brosche, M.; Van Breusegem, F.; Kangasjarvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Van Breusegem, F.; Mhamdi, A. Redox-dependent control of nuclear transcription in plants. J. Exp. Bot. 2018, 69, 3359–3372. [Google Scholar] [CrossRef]
- Muller-Moule, P.; Havaux, M.; Niyogi, K.K. Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol. 2003, 133, 748–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.P.; Muller-Moule, P.; Gilmore, A.M.; Niyogi, K.K. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 2002, 99, 15222–15227. [Google Scholar] [CrossRef] [Green Version]
- Muller-Moule, P.; Golan, T.; Niyogi, K.K. Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 2004, 134, 1163–1172. [Google Scholar] [CrossRef] [Green Version]
- Green, M.A.; Fry, S.C. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-L-threonate. Nature 2005, 433, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obara, K.; Sumi, K.; Fukuda, H. The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol. 2002, 43, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.A.; Nakano, Y.; Asada, K. Monodehydroascorbate reductase in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen peroxide. Plant Cell Physiol. 1984, 25, 385–395. [Google Scholar] [CrossRef]
- Hossain, M.A.; Asada, K. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J. Biol. Chem. 1985, 260, 12920–12926. [Google Scholar] [CrossRef]
- Sano, S.; Miyake, C.; Mikami, B.; Asada, K. Molecular characterization of monodehydroascorbate radical reductase from cucumber highly expressed in Escherichia coli. J. Biol. Chem. 1995, 270, 21354–21361. [Google Scholar] [CrossRef] [Green Version]
- Sano, S.; Tao, S.; Endo, Y.; Inaba, T.; Hossain, M.A.; Miyake, C.; Matsuo, M.; Aoki, H.; Asada, K.; Saito, K. Purification and cDNA cloning of chloroplastic monodehydroascorbate reductase from spinach. Biosci. Biotechnol. Biochem. 2005, 69, 762–772. [Google Scholar] [CrossRef]
- Vanacker, H.; Guichard, M.; Bohrer, A.S.; Issakidis-Bourguet, E. Redox regulation of monodehydroascorbate reductase by thioredoxin y in plastids revealed in the context of water stress. Antioxidants 2018, 7, 183. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Gallie, D.R. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 2004, 16, 1143–1162. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Gallie, D.R. Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 2006, 142, 775–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Gallie, D.R. Dehydroascorbate reductase affects non-photochemical quenching and photosynthetic performance. J. Biol. Chem. 2008, 283, 21347–21361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahantaniaina, M.S.; Li, S.; Chatel-Innocenti, G.; Tuzet, A.; Issakidis-Bourguet, E.; Mhamdi, A.; Noctor, G. Cytosolic and chloroplastic DHARs cooperate in oxidative stress-driven activation of the salicylic acid pathway. Plant Physiol. 2017, 174, 956–971. [Google Scholar] [CrossRef] [Green Version]
- Eastmond, P.J. Monodehyroascorbate reductase4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 2007, 19, 1376–1387. [Google Scholar] [CrossRef] [Green Version]
- Johnston, E.J.; Rylott, E.L.; Beynon, E.; Lorenz, A.; Chechik, V.; Bruce, N.C. Monodehydroascorbate reductase mediates TNT toxicity in plants. Science 2015, 349, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Haroldsen, V.M.; Chi-Ham, C.L.; Kulkarni, S.; Lorence, A.; Bennett, A.B. Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol. Biochem. 2011, 49, 1244–1249. [Google Scholar] [CrossRef] [Green Version]
- Gest, N.; Garchery, C.; Gautier, H.; Jimenez, A.; Stevens, R. Light-dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol. J. 2013, 11, 344–354. [Google Scholar] [CrossRef]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Rio, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Carrie, C.; Law, S.R.; Murcha, M.W.; Whelan, J. Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol. 2013, 161, 644–662. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wu, Q.Y.; Sun, Y.L.; Wang, L.Y.; Yang, X.H.; Meng, Q.W. Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol. Plant 2010, 139, 421–434. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef]
- Li, F.W.; Nishiyama, T.; Waller, M.; Frangedakis, E.; Keller, J.; Li, Z.; Fernandez-Pozo, N.; Barker, M.S.; Bennett, T.; Blazquez, M.A.; et al. Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat. Plants 2020, 6, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996, 12, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kameoka, T.; Okayasu, T.; Kikuraku, K.; Ogawa, T.; Sawa, Y.; Yamamoto, H.; Ishikawa, T.; Maruta, T. Cooperation of chloroplast ascorbate peroxidases and proton gradient regulation 5 is critical for protecting Arabidopsis plants from photooxidative stress. Plant J. 2021, 107, 876–892. [Google Scholar] [CrossRef]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. Oxidative stress and antioxidative systems: Recipes for successful data collection and interpretation. Plant Cell Environ. 2016, 39, 1140–1160. [Google Scholar] [CrossRef] [Green Version]
- Shiroma, S.; Tanaka, M.; Sasaki, T.; Ogawa, T.; Yoshimura, K.; Sawa, Y.; Maruta, T.; Ishikawa, T. Chloroplast development activates the expression of ascorbate biosynthesis-associated genes in Arabidopsis roots. Plant Sci. 2019, 284, 185–191. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara genome: Secondary complexity and implications for plant terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, A.K.; Kim, I.S.; Do, H.; Jeon, B.W.; Lee, C.W.; Roh, S.J.; Shin, S.C.; Park, H.; Kim, Y.S.; Kim, Y.H.; et al. Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica. Sci. Rep. 2016, 6, 33903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Sano, S. Molecular and functional characterization of monodehydro-ascorbate and dehydroascorbate reductases. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Burritt, D.J., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 129–156. [Google Scholar]
- Takeda, T.; Yoshimura, K.; Yoshii, M.; Kanahoshi, H.; Miyasaka, H.; Shigeoka, S. Molecular characterization and physiological role of ascorbate peroxidase from halotolerant Chlamydomonas sp. W80 strain. Arch. Biochem. Biophys. 2000, 376, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Hu, J. Defining the plant peroxisomal proteome: From Arabidopsis to rice. Front. Plant Sci. 2011, 2, 103. [Google Scholar] [CrossRef] [Green Version]
- Lingner, T.; Kataya, A.R.; Antonicelli, G.E.; Benichou, A.; Nilssen, K.; Chen, X.Y.; Siemsen, T.; Morgenstern, B.; Meinicke, P.; Reumann, S. Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 2011, 23, 1556–1572. [Google Scholar] [CrossRef] [Green Version]
- Reumann, S.; Babujee, L.; Ma, C.; Wienkoop, S.; Siemsen, T.; Antonicelli, G.E.; Rasche, N.; Luder, F.; Weckwerth, W.; Jahn, O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 2007, 19, 3170–3193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reumann, S.; Quan, S.; Aung, K.; Yang, P.; Manandhar-Shrestha, K.; Holbrook, D.; Linka, N.; Switzenberg, R.; Wilkerson, C.G.; Weber, A.P.; et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 2009, 150, 125–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Aoki, M.; Kondo, M.; Nishimura, M. Changes in targeting efficiencies of proteins to plant microbodies caused by amino acid substitutions in the carboxy-terminal tripeptide. Plant Cell Physiol. 1997, 38, 759–768. [Google Scholar] [CrossRef] [Green Version]
- Van Norman, J.M.; Frederick, R.L.; Sieburth, L.E. BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr. Biol. 2004, 14, 1739–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Suorsa, M.; Jarvi, S.; Grieco, M.; Nurmi, M.; Pietrzykowska, M.; Rantala, M.; Kangasjarvi, S.; Paakkarinen, V.; Tikkanen, M.; Jansson, S.; et al. Proton gradient regulation5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 2012, 24, 2934–2948. [Google Scholar] [CrossRef] [Green Version]
- Kono, M.; Yamori, W.; Suzuki, Y.; Terashima, I. Photoprotection of PSI by far-red light against the fluctuating light-induced photoinhibition in Arabidopsis thaliana and field-grown plants. Plant Cell Physiol. 2017, 58, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Takagi, D.; Takumi, S.; Hashiguchi, M.; Sejima, T.; Miyake, C. Superoxide and singlet oxygen produced within the thylakoid membranes both cause photosystem I photoinhibition. Plant Physiol. 2016, 171, 1626–1634. [Google Scholar] [CrossRef] [Green Version]
- Yabuta, Y.; Mieda, T.; Rapolu, M.; Nakamura, A.; Motoki, T.; Maruta, T.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Light regulation of ascorbate biosynthesis is dependent on the photosynthetic electron transport chain but independent of sugars in Arabidopsis. J. Exp. Bot. 2007, 58, 2661–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narendra, S.; Venkataramani, S.; Shen, G.; Wang, J.; Pasapula, V.; Lin, Y.; Kornyeyev, D.; Holaday, A.S.; Zhang, H. The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Panchuk, I.I.; Zentgraf, U.; Volkov, R.A. Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 2005, 222, 926–932. [Google Scholar] [CrossRef]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef] [PubMed]
- Rahantaniaina, M.S.; Li, S.; Chatel-Innocenti, G.; Tuzet, A.; Mhamdi, A.; Vanacker, H.; Noctor, G. Glutathione oxidation in response to intracellular H2O2: Key but overlapping roles for dehydroascorbate reductases. Plant Signal. Behav. 2017, 12, e1356531. [Google Scholar] [CrossRef] [Green Version]
- Sha, S.; Minakuchi, K.; Higaki, N.; Sato, K.; Ohtsuki, K.; Kurata, A.; Yoshikawa, H.; Kotaru, M.; Masumura, T.; Ichihara, K.; et al. Purification and characterization of glutaredoxin (thioltransferase) from rice (Oryza sativa L.). J. Biochem. 1997, 121, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Vlamis-Gardikas, A.; Lillig, C.H.; Berndt, C.; Schwenn, J.D.; Holmgren, A.; Jacquot, J.P. Characterization of the redox properties of poplar glutaredoxin. Antioxid. Redox Signal. 2003, 5, 15–22. [Google Scholar] [CrossRef]
- Loi, M.; Leonardis, S.; Mule, G.; Logrieco, A.F.; Paciolla, C. A Novel and potentially multifaceted dehydroascorbate reductase increasing the antioxidant systems is induced by Beauvericinin Tomato. Antioxidants 2020, 9, 435. [Google Scholar] [CrossRef]
- Morell, S.; Follmann, H.; De Tullio, M.; Haberlein, I. Dehydroascorbate and dehydroascorbate reductase are phantom indicators of oxidative stress in plants. FEBS Lett. 1997, 414, 567–570. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, M.; Furutani, R.; Sohtome, T.; Suzuki, T.; Wada, S.; Tanaka, S.; Ifuku, K.; Ueno, D.; Miyake, C. Photosynthetic parameters show specific responses to essential mineral deficiencies. Antioxidants 2020, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Little, D.; Gouhier-Darimont, C.; Bruessow, F.; Reymond, P. Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiol. 2007, 143, 784–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, M.; Takahashi, R.; Hamada, A.; Terai, Y.; Ogawa, T.; Sawa, Y.; Ishikawa, T.; Maruta, T. Distribution and Functions of Monodehydroascorbate Reductases in Plants: Comprehensive Reverse Genetic Analysis of Arabidopsis thaliana Enzymes. Antioxidants 2021, 10, 1726. https://doi.org/10.3390/antiox10111726
Tanaka M, Takahashi R, Hamada A, Terai Y, Ogawa T, Sawa Y, Ishikawa T, Maruta T. Distribution and Functions of Monodehydroascorbate Reductases in Plants: Comprehensive Reverse Genetic Analysis of Arabidopsis thaliana Enzymes. Antioxidants. 2021; 10(11):1726. https://doi.org/10.3390/antiox10111726
Chicago/Turabian StyleTanaka, Mio, Ryuki Takahashi, Akane Hamada, Yusuke Terai, Takahisa Ogawa, Yoshihiro Sawa, Takahiro Ishikawa, and Takanori Maruta. 2021. "Distribution and Functions of Monodehydroascorbate Reductases in Plants: Comprehensive Reverse Genetic Analysis of Arabidopsis thaliana Enzymes" Antioxidants 10, no. 11: 1726. https://doi.org/10.3390/antiox10111726