Distinct H3K27me3 and H3K27ac Modifications in Neural Tube Defects Induced by Benzo[a]pyrene
Abstract
:1. Introduction
2. Materials and Methods
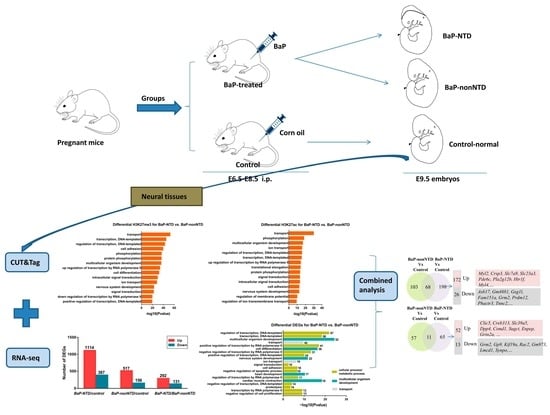
2.1. Animal Experiments
2.2. Isolation of Neural Tissues
2.3. RNA-seq
2.4. Analysis of Differential Gene Expression and Functional Annotation
2.5. CUT&Tag Experiment
2.6. CUT&Tag Data Processing and Analysis
2.7. Quantitative Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR)
2.8. Statistical Analyses
3. Results
3.1. CUT&Tag Analysis
3.2. Transcriptome Analysis
3.3. Combined Analysis of Histone Modifications and Transcription
3.4. Motif Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greene, N.D.; Copp, A.J. Neural tube defects. Annu. Rev. Neurosci. 2014, 37, 221–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christianson, A.; Howson, C.; Modell, C. March of Dimes Global Report on Birth Defects: The Hidden Toll of Dying and Disabled Children. White Plains, New York: March Dimes Birth Defects Found. 2006. Available online: https://weekly.chinacdc.cn/fileCCDCW/journal/article/ccdcw/2020/37/PDF/CCDCW200137.pdf (accessed on 1 September 2020).
- World Health Organization. Global Health Estimates (GHE)–Cause-Specific Mortality. 2015. Available online: http://www.who.int/healthinfo/global_burden_disease/estimates/en/index1.html (accessed on 14 April 2015).
- World Health Organization. Global Health Estimates (GHE)–Disease Burden. 2015. Available online: http://www.who.int/healthinfo/global_burden_disease/estimates/en/index2.html (accessed on 14 April 2015).
- Blencowe, H.; Cousens, S.; Modell, B.; Lawn, J. Folic acid to reduce neonatal mortality from neural tube disorders. Int. J. Epidemiol. 2010, 39 (Suppl. 1), i110–i121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, N.D.; Copp, A.J. Inositol prevents folate-resistant neural tube defects in the mouse. Nat. Med. 1997, 3, 60–66. [Google Scholar] [CrossRef] [PubMed]
- MRC Vitamin Study Research Group. Prevention of neural tube defects: Results of the Medical Research Council Vitamin Study. Lancet 1991, 338, 131–137. [Google Scholar] [CrossRef]
- Harris, M.J.; Juriloff, D.M. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res. Part A Clin. Mol. Teratol. 2010, 88, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Stanier, P.; DE Greene, N. Neural tube defects: Recent advances, unsolved questions, and controversies. Lancet Neurol. 2013, 12, 799–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Ren, A.; Wang, L.; Huang, Y.; Wang, Y.; Wang, C.; Greene, N.D. Oxidative Stress and Apoptosis in Benzo[a]pyrene-Induced Neural Tube Defects. Free Radic. Biol. Med. 2018, 116, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jin, L.; Ren, A.; Yuan, Y.; Liu, J.; Li, Z.; Zhang, L.; Yi, D.; Wang, L.-L.; Zhang, Y.; et al. Levels of polycyclic aromatic hydrocarbons in maternal serum and risk of neural tube defects in offspring. Environ. Sci. Technol. 2015, 49, 588–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, B.; Chen, S.; Zhang, Q.; Jiang, Q.; Li, H. Abnormal epigenetic regulation of the gene expression levels of Wnt2b and Wnt7b: Implications for neural tube defects. Mol. Med. Rep. 2016, 13, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Bai, B.; Zhang, Q.; Liu, X.; Miao, C.; Shangguan, S.; Bao, Y.; Guo, J.; Wang, L.; Zhang, T.; Li, H. Different epigenetic alterations are associated with abnormal IGF2/Igf2 upregulation in neural tube defects. PLoS ONE 2014, 9, e113308. [Google Scholar] [CrossRef] [Green Version]
- Wan, C.; Liu, X.; Bai, B.; Cao, H.; Li, H.; Zhang, Q. Regulation of the expression of tumor necrosis factor-related genes by abnormal histone H3K27 acetylation: Implications for neural tube defects. Mol. Med. Rep. 2018, 17, 8031–8038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Wu, Y.; Yang, P. High glucose-induced oxidative stress represses sirtuin deacetylase expression and increases histone acetylation leading to neural tube defects. J. Neurochem. 2016, 137, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Xue, P.; Li, H.; Bao, Y.; Wu, L.; Chang, S.; Niu, B.; Yang, F.; Zhang, T. Histone modification mapping in human brain reveals aberrant expression of histone H3 lysine 79 dimethylation in neural tube defects. Neurobiol. Dis. 2013, 54, 404–413. [Google Scholar] [CrossRef]
- Zhang, Q.; Bai, B.; Mei, X.; Wan, C.; Cao, H.; Li, D.; Wang, S.; Zhang, M.; Wang, Z.; Wu, J.; et al. Elevated H3K79 homocysteinylation causes abnormal gene expression during neural development and subsequent neural tube defects. Nat. Commun. 2018, 9, 3436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 193. [Google Scholar] [CrossRef] [Green Version]
- Niu, W.; Zou, Y.; Shen, C.; Zhang, C.-L. Activation of Postnatal Neural Stem Cells Requires Nuclear Receptor TLX. J. Neurosci. 2011, 31, 13816–13828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, P.; Brock, G.; Appana, S.; Webb, C.; Greene, R.M.; Pisano, M.M. MicroRNA gene expression signatures in the developing neural tube. Birth Defects Res. Part A Clin. Mol. Teratol. 2011, 91, 744–762. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- The Gene Ontology Project in 2008. Nucleic Acids Res. 2008, 36, D440–D444. [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, Y.H.; Johnson, T.D.; Rozek, L.S.; Sartor, M.A. PePr: A peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics 2014, 30, 2568–2575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langlois, P.H.; Hoyt, A.T.; Lupo, P.; Lawson, C.C.; Waters, M.A.; Desrosiers, T.A.; Shaw, G.M.; Romitti, P.A.; Lammer, E.J. The National Birth Defects Prevention Study Maternal occupational exposure to polycyclic aromatic hydrocarbons and risk of neural tube defect-affected pregnancies. Birth Defects Res. Part A Clin. Mol. Teratol. 2012, 94, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copp, A.J. Neurulation in the cranial region—normal and abnormal. J. Anat. 2005, 207, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.D.E.; Copp, A.J. Development of the vertebrate central nervous system: Formation of the neural tube. Prenat. Diagn. 2009, 29, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.G.A., 4th; Uribe, R.A. Hox proteins as regulators of extracellular matrix interactions during neural crest migration. Differentiation 2022, 128, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.D.; Copp, A.J. Mouse models of neural tube defects: Investigating preventive mechanisms. Am. J. Med. Genet. Part C Semin. Med. Genet. 2005, 135C, 31–41. [Google Scholar] [CrossRef]
- Stolfi, A.; Wagner, E.; Taliaferro, J.M.; Chou, S.; Levine, M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development 2011, 138, 5429–5439. [Google Scholar] [CrossRef] [Green Version]
- Copp, A.J.; Greene, N.D.E. Neural tube defects-disorders of neurulation and related embryonic processes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Goyal, R.; Spencer, K.A.; Borodinsky, L.N. From Neural Tube Formation Through the Differentiation of Spinal Cord Neurons: Ion Channels in Action During Neural Development. Front. Mol. Neurosci. 2020, 13, 62. [Google Scholar] [CrossRef]
- Patterson, E.S.; Waller, L.E.; Kroll, K.L. Geminin loss causes neural tube defects through disrupted progenitor specification and neuronal differentiation. Dev. Biol. 2014, 393, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhou, W.; Li, X.; Gao, M.; Ji, S.; Tian, W.; Ji, G.; Du, J.; Hao, A. SOX19b regulates the premature neuronal differentiation of neural stem cells through EZH2-mediated histone methylation in neural tube development of zebrafish. Stem Cell Res. Ther. 2019, 10, 389. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ji, G.; Zhou, J.; Du, J.; Li, X.; Shi, W.; Hu, Y.; Zhou, W.; Hao, A. Pcgf1 Regulates Early Neural Tube Development Through Histone Methylation in Zebrafish. Front. Cell Dev. Biol. 2020, 8, 581636. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, S.; Allen, M.; Cox, C.L.; Walker, L.P.; Malphrus, K.; Qiu, Y.; Nguyen, T.; Rowley, C.; Kouri, N.; Crook, J.; et al. LRRTM3 Interacts with APP and BACE1 and Has Variants Associating with Late-Onset Alzheimer’s Disease (LOAD). PLoS ONE 2013, 8, e64164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darsalia, V.; Ortsäter, H.; Olverling, A.; Darlöf, E.; Wolbert, P.; Nyström, T.; Klein, T.; Sjöholm, Å.; Patrone, C. The DPP-4 inhibitor linagliptin counteracts stroke in the normal and diabetic mouse brain: A comparison with glimepiride. Diabetes 2013, 62, 1289–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, S.V.; Ward, J.M.; Tessarollo, L.; McAreavey, D.; Sachdev, V.; Fananapazir, L.; Banks, M.K.; Morris, N.; Djurickovic, D.; Devor-Henneman, D.E.; et al. Cerebellar Ataxia, Seizures, Premature Death, and Cardiac Abnormalities in Mice with Targeted Disruption of the Cacna2d2 Gene. Am. J. Pathol. 2004, 165, 1007–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, B.; D’Andrea, D.; Collins, M.O.; Rees, E.; Steward, T.G.J.; Zhu, Y.; Chapman, G.; Legge, S.E.; Pardiñas, A.F.; Harwood, A.J.; et al. Transcriptional programs regulating neuronal differentiation are disrupted in DLG2 knockout human embryonic stem cells and enriched for schizophrenia and related disorders risk variants. Nat. Commun. 2022, 13, 27. [Google Scholar] [CrossRef]
- Chakravarti, B.; Yang, J.; Ahlers-Dannen, K.E.; Luo, Z.; Flaherty, H.A.; Meyerholz, D.K.; Anderson, M.E.; Fisher, R.A. Essentiality of Regulator of G Protein Signaling 6 and Oxidized Ca(2+)/Calmodulin-Dependent Protein Kinase II in Notch Signaling and Cardiovascular Development. J. Am. Heart Assoc. 2017, 6, e007038. [Google Scholar] [CrossRef] [Green Version]
- Amarnath, S.; Agarwala, S. Cell cycle dependent TGFβ-BMP antagonism regulates neural tube closure by modulating tight junctions. J. Cell Sci. 2017, 130, 119–131. [Google Scholar] [CrossRef] [Green Version]
- LeSueur, J.A.; Fortuno, E.S., 3rd; McKay, R.M.; Graff, J.M. Smad10 is required for formation of the frog nervous system. Dev. Cell 2002, 2, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Wang, R.; Li, X.; Yu, L.; Hua, D.; Sun, C.; Shi, C.; Luo, W.; Rao, C.; Jiang, Z.; et al. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J. Clin. Investig. 2019, 129, 676–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anczuków, O.; Akerman, M.; Cléry, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comiskey, D.F.; Jacob, A.G.; Singh, R.K.; Tapia-Santos, A.S.; Chandler, D.S. Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res. 2015, 43, 4202–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Zhang, W.; Zhao, J.; Sun, B.; Qi, Y.; Ji, H.; Chen, C.; Zhang, J.; Sheng, J.; Wang, T.; et al. SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer. Signal Transduct. Target. Ther. 2021, 6, 108. [Google Scholar] [CrossRef]
- Jiang, M.; Feng, J.; Fu, R.; Pan, Y.; Liu, X.; Dai, J.; Jiang, C.; Hao, Y.; Ren, M. Transfection of STAT3 overexpression plasmid mediated through recombinant lentivirus promotes differentiation of bone marrow mesenchymal stem cells into neural cells in fetal rats with spina bifida aperta. Aging 2021, 13, 21778–21790. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Niswander, L. Zinc deficiency causes neural tube defects through attenuation of p53 ubiquitylation. Development 2018, 145, dev169797. [Google Scholar] [CrossRef] [Green Version]
- Shirane, M.; Ogawa, M.; Motoyama, J.; Nakayama, K.I. Regulation of apoptosis and neurite extension by FKBP38 is required for neural tube formation in the mouse. Genes Cells 2008, 13, 635–651. [Google Scholar] [CrossRef]
- Steber, C.M.; E Esposito, R. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA 1995, 92, 12490–12494. [Google Scholar] [CrossRef] [Green Version]
- Strich, R.; Surosky, R.T.; Steber, C.; Dubois, E.; Messenguy, F.; E Esposito, R. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994, 8, 796–810. [Google Scholar] [CrossRef] [Green Version]
- Bartholomew, C.R.; Suzuki, T.; Du, Z.; Backues, S.K.; Jin, M.; Lynch-Day, M.A.; Umekawa, M.; Kamath, A.; Zhao, M.; Xie, Z.; et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. USA 2012, 109, 11206–11210. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.-Y.; He, T.-T.; Gao, X.-M.; Zhao, Y.; Wang, J. ZBTB Transcription Factors: Key Regulators of the Development, Differentiation and Effector Function of T Cells. Front. Immunol. 2021, 12, 713294. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, D.; Wang, T.; He, S. Pyrimidine Biosynthetic Enzyme CAD: Its Function, Regulation, and Diagnostic Potential. Int. J. Mol. Sci. 2021, 22, 10253. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Vuong, A.M.; Dietrich, K.N.; Chen, A. Proximity to traffic and exposure to polycyclic aromatic hydrocarbons in relation to Attention Deficit Hyperactivity Disorder and conduct disorder in U.S. children. Int. J. Hyg. Environ. Health. 2021, 232, 113686. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kalia, V.; Perera, F.; Herbstman, J.; Li, T.; Nie, J.; Qu, L.; Yu, J.; Tang, D. Prenatal airborne polycyclic aromatic hydrocarbon exposure, LINE1 methylation and child development in a Chinese cohort. Environ. Int. 2017, 99, 315–320. [Google Scholar] [CrossRef]
- Perera, F.P.; Rauh, V.; Whyatt, R.M.; Tsai, W.Y.; Tang, D.; Diaz, D.; Hoepner, L.; Barr, D.; Tu, Y.-H.; Camann, D.; et al. Effect of prenatal exposure to airborne polycyclic aromatic hydrocarbons on neurodevelopment in the first 3 years of life among inner-city children. Environ Health Perspect 2006, 114, 1287–1292. [Google Scholar] [CrossRef] [Green Version]
- Allen, J.L.; Klocke, C.; Morris-Schaffer, K.; Conrad, K.; Sobolewski, M.; Cory-Slechta, D.A. Cognitive Effects of Air Pollution Exposures and Potential Mechanistic Underpinnings. Curr. Environ. Health. Rep. 2017, 4, 180–191. [Google Scholar] [CrossRef]
- Legraverend, C.; Guenthner, T.M.; Nebert, D.W. Importance of the route of administration for genetic differences in benzo[a]pyrene-induced in utero toxicity and teratogenicity. Teratology 1984, 29, 35–47. [Google Scholar] [CrossRef]
- Nicol, C.J.; Harrison, M.L.; Laposa, R.R.; Gimelshtein, I.L.; Wells, P.G. A teratologic suppressor role for p53 in benzo(a)pyrene–treated transgenic p53-deficient mice. Nat. Genet. 1995, 10, 181–187. [Google Scholar] [CrossRef]
- Lin, S.; Ren, A.; Wang, L.; Santos, C.; Huang, Y.; Jin, L.; Li, Z.; Greene, N.D.E. Aberrant methylation of Pax3 gene and neural tube defects in association with exposure to polycyclic aromatic hydrocarbons. Clin. Epigenetics 2019, 11, 13. [Google Scholar] [CrossRef]





| Gene ID | Gene Name | Expression FC | Expression State | Histone Modification | Modification State | Location |
|---|---|---|---|---|---|---|
| 17906 | Myl2 | 6.787 | Up-regulated | H3K27ac | Decreased | Genebody |
| 383348 | Kctd16 | 2.997 | Up-regulated | H3K27ac | Increased | Genebody |
| 65256 | Asb2 | 2.159 | Up-regulated | H3K27me3 | Increased | Genebody |
| 213435 | Mylk3 | 2.084 | Up-regulated | H3K27me3 | Decreased | Genebody |
| 19415 | Rasal1 | 2.016 | Up-regulated | H3K27me3 | Increased | Genebody |
| 13482 | Dpp4 | 1.603 | Up-regulated | H3K27me3 | Decreased | Promoter |
| 270049 | Galntl6 | 1.484 | Up-regulated | H3K27me3 | Decreased | Genebody |
| 23859 | Dlg2 | 1.47 | Up-regulated | H3K27me3 | Decreased | Genebody |
| 14799 | Gria1 | 1.47 | Up-regulated | H3K27me3 | Increased | Genebody |
| 74189 | Phactr3 | 1.454 | Down-regulated | H3K27me3 | Decreased | Genebody |
| 216033 | Ctnna3 | 1.428 | Up-regulated | H3K27me3 | Increased | Genebody |
| 545428 | Ccdc141 | 1.269 | Up-regulated | H3K27ac | Decreased | Genebody |
| 56808 | Cacna2d2 | 1.247 | Up-regulated | H3K27me3 | Decreased | Genebody |
| 50779 | Rgs6 | 1.246 | Up-regulated | H3K27me3 | Increased | Genebody |
| 50787 | Hs6st3 | 1.154 | Up-regulated | H3K27ac | Decreased | Genebody |
| 237558 | Myrfl | 1.088 | Up-regulated | H3K27me3 | Decreased | Genebody |
| 22041 | Trf | 1.083 | Up-regulated | H3K27me3 | Increased | Genebody |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Wang, C.; Li, Z.; Qiu, X. Distinct H3K27me3 and H3K27ac Modifications in Neural Tube Defects Induced by Benzo[a]pyrene. Brain Sci. 2023, 13, 334. https://doi.org/10.3390/brainsci13020334
Lin S, Wang C, Li Z, Qiu X. Distinct H3K27me3 and H3K27ac Modifications in Neural Tube Defects Induced by Benzo[a]pyrene. Brain Sciences. 2023; 13(2):334. https://doi.org/10.3390/brainsci13020334
Chicago/Turabian StyleLin, Shanshan, Chengrui Wang, Zhiwen Li, and Xiu Qiu. 2023. "Distinct H3K27me3 and H3K27ac Modifications in Neural Tube Defects Induced by Benzo[a]pyrene" Brain Sciences 13, no. 2: 334. https://doi.org/10.3390/brainsci13020334