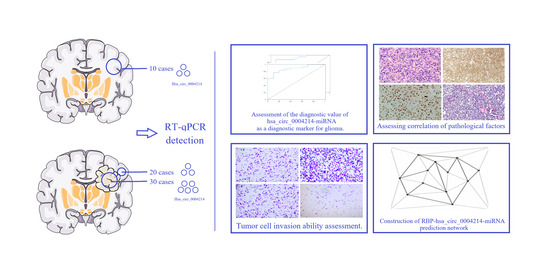
A Meaningful Strategy for Glioma Diagnosis via Independent Determination of hsa_circ_0004214
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Tissue Specimens and Ethical Approval
2.2. Cell Culture and Transfection
2.3. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.4. Fluorescence In Situ Hybridization (FISH)
2.5. Transwell Migration Assay
2.6. Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
3.1. hsa_circ_0004214 Stable Expression in Glioma
3.2. High hsa_circ_0004214 Expression in Human Glioma Tissue
3.3. Expression of hsa_circ_0004214 in Different Grades of Gliomas
3.4. Diagnostic Value of hsa_circ_0004214
3.5. Relationship of hsa_circ_0004214 Levels with Clinicopathological Factors of Patients with Glioma
3.6. hsa_circ_0004214 Affects the Migration Capacity of Glioma In Vitro
3.7. Construction of Regulatory Network Associated with hsa_circ_0004214
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, L.; Wang, Q.; Shen, M.; Sun, Y.; Zhang, X.; Huang, C.; Chen, J.; Li, R.; Duan, Y. Thermoresponsive nanocomposite gel for local drug delivery to suppress the growth of glioma by inducing autophagy. Autophagy 2017, 13, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Vannini, E.; Mori, E.; Tantillo, E.; Schmidt, G.; Caleo, M.; Costa, M. CTX-CNF1 Recombinant Protein Selectively Targets Glioma Cells In Vivo. Toxins 2021, 13, 194. [Google Scholar] [CrossRef]
- Cheng, M.; Zhang, Z.W.; Ji, X.H.; Xu, Y.; Bian, E.; Zhao, B. Super-enhancers: A new frontier for glioma treatment. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1873, 188353. [Google Scholar] [CrossRef] [PubMed]
- Mondal, I.; Kulshreshtha, R. Potential of microRNA based diagnostics and therapeutics in glioma: A patent review. Expert Opin. Ther. Patents 2020, 31, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, H.; Pan, J.; Yan, T.; Zhou, H.; Han, X.; Su, L.; Hou, L.; Xue, X. Upregulated Expression of Cancer-Derived Immunoglobulin G Is Associated with Progression in Glioma. Front. Oncol. 2021, 11, 758856. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liang, R.; Song, C.; Xiang, Y.; Liu, Y. Prognostic and clinicopathological significance of long non-coding RNA in glioma. Neurosurg. Rev. 2018, 43, 1–8. [Google Scholar] [CrossRef]
- Sun, W.; Zhou, H.; Han, X.; Hou, L.; Xue, X. Circular RNA: A novel type of biomarker for glioma. Mol. Med. Rep. 2021, 24, 1–12. [Google Scholar] [CrossRef]
- Peng, L.; Sang, H.; Wei, S.; Li, Y.; Jin, D.; Zhu, X.; Li, X.; Dang, Y.; Zhang, G. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol. Cancer 2020, 19, 156. [Google Scholar] [CrossRef]
- Li, P.; Xu, Z.; Liu, T.; Liu, Q.; Zhou, H.; Meng, S.; Feng, Z.; Tang, Y.; Liu, C.; Feng, J.; et al. Circular RNA Sequencing Reveals Serum Exosome Circular RNA Panel for High-Grade Astrocytoma Diagnosis. Clin. Chem. 2022, 68, 332–343. [Google Scholar] [CrossRef]
- de Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, B.; Klinge, C.M. Regulation of breast cancer metastasis signaling by miRNAs. Cancer Metastasis Rev. 2020, 39, 837–886. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, H.; Li, Z.; Li, F.; Liang, L.; Zou, Y.; Shen, H.; Li, J.; Xia, Y.; Cheng, Z.; et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 2022, 76, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2018, 98, 87–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Wu, Z.; Zhang, H.; Zhang, N.; Wu, W.; Wang, Z.; Dai, Z.; Zhang, X.; Zhang, L.; Peng, Y.; et al. Glioma targeted therapy: Insight into future of molecular approaches. Mol. Cancer. 2022, 21, 39. [Google Scholar] [CrossRef]
- Bagley, S.J.; Kothari, S.; Rahman, R.; Lee, E.Q.; Dunn, G.P.; Galanis, E.; Chang, S.M.; Nabors, L.B.; Ahluwalia, M.S.; Stupp, R.; et al. Glioblastoma Clinical Trials: Current Landscape and Opportunities for Improvement. Clin. Cancer Res. 2021, 28, 594–602. [Google Scholar] [CrossRef]
- Liu, J.; Yang, Q.; Sun, H.; Wang, X.; Saiyin, H.; Zhang, H. The circ-AMOTL1/ENO1 Axis Implicated in the Tumorigenesis of OLP-Associated Oral Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 7219–7230. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Z.; Zhou, J.; Tian, C.; Tian, G.; He, M.; Gao, L.; Chen, L.; Li, T.; Peng, H.; et al. Interior circular RNA. RNA Biol. 2019, 17, 87–97. [Google Scholar] [CrossRef]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 417. [Google Scholar] [CrossRef] [Green Version]
- Ou, R.; Lv, J.; Zhang, Q.; Lin, F.; Zhu, L.; Huang, F.; Li, X.; Li, T.; Zhao, L.; Ren, Y.; et al. circAMOTL1 Motivates AMOTL1 Expression to Facilitate Cervical Cancer Growth. Mol. Ther.-Nucleic Acids 2019, 19, 50–60. [Google Scholar] [CrossRef]
- Zhu, Z.; Rong, Z.; Luo, Z.; Yu, Z.; Zhang, J.; Qiu, Z.; Huang, C. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol. Cancer 2019, 18, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; He, Y.-R.; Liang, L.-C.; Huang, Q.; Zhu, Z.-Q. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J. Gastroenterol. 2017, 23, 6330–6338. [Google Scholar] [CrossRef] [PubMed]
- van Bodegraven, E.J.; van Asperen, J.V.; Robe, P.A.J.; Hol, E.M. Importance of GFAP isoform-specific analyses in astrocytoma. Glia 2019, 67, 1417–1433. [Google Scholar] [CrossRef] [PubMed]
- Perry, A.; Burton, S.S.; Fuller, G.N.; Robinson, C.A.; Palmer, C.A.; Resch, L.; Bigio, E.H.; Gujrati, M.; Rosenblum, M.K. Oligodendroglial neoplasms with ganglioglioma-like maturation: A diagnostic pitfall. Acta Neuropathol. 2010, 120, 237–252. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, A.S.; Rosa, M.D.C.; Borodovsky, A.; Festuccia, W.; Chan, T.; Riggins, G.J. Demethylation and epigenetic modification with 5-azacytidine reduces IDH1 mutant glioma growth in combination with temozolomide. Neuro-Oncology 2018, 21, 189–200. [Google Scholar] [CrossRef]
- Liu, S.; Zhu, Y.; Zhang, C.; Meng, X.; Sun, B.; Zhang, G.; Fan, Y.; Kang, X. The Clinical Significance of Soluble Programmed Cell Death-Ligand 1 (sPD-L1) in Patients with Gliomas. Front. Oncol. 2020, 10, 9. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Qin, H.; Cheung, F.K.M.; Su, J.; Zhang, D.-D.; Liu, S.-Y.; Li, X.-F.; Qin, J.; Lin, J.-T.; Jiang, B.-Y.; et al. Plasma extracellular vesicle microRNAs for pulmonary ground-glass nodules. J. Extracell. Vesicles 2019, 8, 1663666. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jiang, D.; Li, W.; Xiang, X.; Zhao, J.; Yu, B.; Wang, C.; He, Z.; Zhu, L.; Yang, Y. Evaluation of serum extracellular vesicles as noninvasive diagnostic markers of glioma. Theranostics 2019, 9, 5347–5358. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, X.; Geng, X.; Shi, L.; Li, Q.; Liu, F.; Fang, C.; Wang, H. Advances in circular RNAs and their role in glioma. Int. J. Oncol. 2020, 57, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Gao, J.; Zheng, R.; Yu, M.; Ren, Y.; Yan, T.; Huang, Y.; Li, Y. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Disease 2020, 11, 123. [Google Scholar] [CrossRef]
- Wang, P.F.; Meng, Z.; Song, H.W.; Yao, K.; Duan, Z.J.; Yu, C.J.; Li, S.W.; Yan, C.X. Preoperative Changes in Hematological Markers and Predictors of Glioma Grade and Survival. Front. Pharmacol. 2018, 9, 886. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, J.; Zhao, J.; Hou, D.; Zhang, H.; Li, L.; Zou, D.; Hu, J.; Zhang, Y.; Jing, Z. MiR-18a-downregulated RORA inhibits the proliferation and tumorigenesis of glioma using the TNF-α-mediated NF-κB signaling pathway. eBioMedicine 2020, 52, 102651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jassam, S.A.; Maherally, Z.; Smith, J.R.; Ashkan, K.; Roncaroli, F.; Fillmore, H.L.; Pilkington, G.J. TNF-α enhancement of CD62E mediates adhesion of non-small cell lung cancer cells to brain endothelium via CD15 in lung-brain metastasis. Neuro-Oncology 2016, 18, 679–690. [Google Scholar] [CrossRef] [Green Version]
- Xiong, S.; Peng, H.; Ding, X.; Wang, X.; Wang, L.; Wu, C.; Wang, S.; Xu, H.; Liu, Y. Circular RNA Expression Profiling and the Potential Role of hsa_circ_0089172 in Hashimoto’s Thyroiditis via Sponging miR125a-3p. Mol. Ther.-Nucleic Acids 2019, 17, 38–48. [Google Scholar] [CrossRef] [PubMed]





| Target ID | Type | Direction | Sequence (5′→3′) | Product Length |
|---|---|---|---|---|
| hsa_circ_0004214 | ①Divergent | Forward primer | GTGAGAACAGATGTGGCCGT | 138 |
| Reverse primer | GCATCCTCGGAACCTCTCATT | |||
| ②Convergent | Forward primer | AGCCCATCCTACAAACAA | 218 | |
| GAPDH | Reverse primer | CCTGAAGAACTGCGACTG | ||
| Forward primer | CTCGCTTCGGCAGCACA | 122 | ||
| Reverse primer | AACGCTTCACGAATTTGCGT |
| Target ID | Official Symbol | Primary Source | Gene Type | Organism | Location |
|---|---|---|---|---|---|
| Angiomotin like 1 | AMOTL1 | Nucleotide: NM_130847 | protein coding | Homo sapiens | 11q21 |
| hsa_circ_0004214 | CircAMOTL1 | GEO: GSE146463 | circ-RNA | hsa | chr11:94532555-94533477(+) |
| Group | n | Mean + SD(∆∆CT) | Specificity | Sensitivity | Youden Index | OR | RR |
|---|---|---|---|---|---|---|---|
| Pericarcinomatous and glioam tissue | 20 | −1.24 ± 2.39 | 0.8 | 0.8 | 0.6 | 16 | 4 |
| Control and glioma | 10 | −1.33 ± 2.35 | 0.9 | 0.7 | 0.6 | 21 | 7 |
| Contor-WHO II and WHO III/IV | 40 | −1.75 ± 1.81 | 0.933 | 0.88 | 0.813 | 120.667 | 13.2 |
| The Title | Goal | AUC | Standard Error (SE) | p | 95% CI | Best Threshold | Sensitivity | Specificity | Cut-Off | AUC Difference | Z | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0004214 level | glioma | 0.880 | 0.055 | 0 < 0.05 | 0.773~0.987 | 0.667 | 0.767 | 0.767 | −14.990 | 0.0510 | 0.7564 | 0.4494 |
| hsa_circ_0004214 level | WHOIII/IV | 0.931 | 0.039 | 0 < 0.05 | 0.855~1.006 | 0.813 | 0.880 | 0.933 | −14.950 |
| Clinicopathological Factor | n | Positive | Negative | Tissue hsa_circ_0004214 | ||
|---|---|---|---|---|---|---|
| Mean ± SD | p Value | |||||
| Age (yr) | <60 | 16 | 11 | 5 | 13.13 ± 1.95 | 0.5135 |
| ≥60 | 14 | 11 | 3 | 13.67 ± 2.48 | ||
| Sex | Male | 16 | 14 | 2 | 12.75 ± 1.57 | 0.0795 |
| Female | 14 | 8 | 6 | 14.18 ± 2.65 | ||
| Male height (cm) | <170 | 5 | 5 | 0 | 13.04 ± 1.30 | 0.2930 |
| ≥170 | 11 | 9 | 2 | 12.12 ± 2.07 | ||
| Female height (cm) | <160 | 4 | 2 | 2 | 14.13 ± 2.72 | 0.9112 |
| ≥160 | 10 | 6 | 4 | 14.31 ± 2.86 | ||
| BMI | 18.5–23.9 | 22 | 17 | 5 | 13.12 ± 2.45 | 0.2307 |
| >23.9 | 8 | 5 | 3 | 14.24 ± 1.22 | ||
| <18.5 | 0 | 0 | 0 | - | ||
| WHO grade | II | 5 | 0 | 5 | 16.31 ± 1.74 | 0.0382(II:III) |
| III | 10 | 7 | 3 | 13.99 ± 1.85 | 0.0001(II:IV) | |
| IV | 15 | 15 | 0 | 12.17 ± 1.42 | 0.0281(III:IV) | |
| Diameter in (cm) | <4 | 12 | 7 | 5 | 13.99 ± 2.75 | 0.2552 |
| ≥4 | 18 | 15 | 3 | 13.04 ± 1.78 | ||
| edema belt (cm) | <1 | 14 | 6 | 8 | 14.74 ± 2.23 | 0.0014 |
| ≥1 | 16 | 16 | 0 | 12.34 ± 1.46 | ||
| Position | Frontal lobe | 12 | 7 | 5 | 13.99 ± 2.42 | 0.8016(F:T) |
| Temporal lobe | 12 | 9 | 3 | 13.42 ± 2.35 | 0.3658(F:O) | |
| Other position | 6 | 6 | 0 | 12.48 ± 1.05 | 0.6700(T:O) | |
| pathological type | astrocytic glioma | 14 | 8 | 6 | 14.24 ± 2.20 | 0.7182(A:O) |
| glioblastoma | 13 | 13 | 0 | 12.22 ± 2.19 | 0.0308(G:A) | |
| Other type | 3 | 1 | 2 | 14.97 ± 2.65 | 0.0595(G:O) | |
| Blood Pressure (mmHg) | <140 | 16 | 13 | 3 | 12.79 ± 2.12 | 0.0721 |
| ≥140 | 14 | 9 | 5 | 14.23 ± 2.08 | ||
| Items | n | Positive | Negative | Tissue hsa_circ_0004214 | ||
|---|---|---|---|---|---|---|
| Mean ± SD | p Value | |||||
| immunohistochemical | Vimentin (+) | 28 | 20 | 8 | 13.48 ± 2.22 | - |
| Vimentin (−) | 0 | 0 | 0 | - | ||
| s-100 (+) | 23 | 19 | 4 | 13.05 ± 1.66 | 0.0586 | |
| s-100 (−) | 7 | 3 | 4 | 14.83 ± 3.21 | ||
| GFAP (+) | 24 | 22 | 2 | 12.86 ± 1.75 | 0.0002 | |
| GFAP (−) | 6 | 0 | 6 | 16.23 ± 1.66 | ||
| SYN (+) | 25 | 19 | 6 | 13.25 ± 2.28 | 0.1283 | |
| SYN (−) | 5 | 3 | 2 | 14.90 ± 0.93 | ||
| EMA (+) | 14 | 13 | 1 | 12.64 ± 1.78 | 0.2962 | |
| EMA (−) | 11 | 8 | 3 | 13.45 ± 1.96 | ||
| CD34 (+) | 17 | 14 | 3 | 12.84 ± 1.81 | 0.9391 | |
| CD34 (−) | 9 | 7 | 2 | 13.51 ± 2.02 | ||
| Ki67 (+) | 22 | 19 | 3 | 12.71 ± 1.77 | 0.0008 | |
| Ki67 (−) | 8 | 3 | 5 | 15.53 ± 1.96 | ||
| Genetics | IDH mutation (+) | 11 | 6 | 5 | 14.42 ± 1.86 | 0.0779 |
| IDH mutation (−) | 19 | 16 | 3 | 12.97 ± 2.22 | ||
| MEMG methylation (+) | 6 | 5 | 1 | 13.92 ± 1.65 | 0.6040 | |
| MEMG methylation (−) | 24 | 17 | 7 | 13.40 ± 2.31 | ||
| 1p/19q lose (+) | 0 | 0 | 0 | - | - | |
| 1p/19q lose (−) | 30 | 22 | 8 | 13.46 ± 2.19 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Zhang, Y.; Tian, J.; Miao, Z.; Lv, S.; Zhao, X. A Meaningful Strategy for Glioma Diagnosis via Independent Determination of hsa_circ_0004214. Brain Sci. 2023, 13, 193. https://doi.org/10.3390/brainsci13020193
Zhou Y, Zhang Y, Tian J, Miao Z, Lv S, Zhao X. A Meaningful Strategy for Glioma Diagnosis via Independent Determination of hsa_circ_0004214. Brain Sciences. 2023; 13(2):193. https://doi.org/10.3390/brainsci13020193
Chicago/Turabian StyleZhou, Yinan, Yating Zhang, Jiajia Tian, Zengli Miao, Shangrui Lv, and Xudong Zhao. 2023. "A Meaningful Strategy for Glioma Diagnosis via Independent Determination of hsa_circ_0004214" Brain Sciences 13, no. 2: 193. https://doi.org/10.3390/brainsci13020193