Combining High-Resolution Hard X-ray Tomography and Histology for Stem Cell-Mediated Distraction Osteogenesis
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Stem Cell Preparation
2.2. Surgical Procedure, Osteotomy and Distractor Application
2.3. Conventional Microtomography
2.4. Decalcification and Paraffin-Embedding of the Mandibles
2.5. Synchrotron Radiation-Based Tomography
2.6. Three-Dimensional Visualization of the Tomography Data
2.7. Histology and Immune-Histochemical Preparation
2.8. Registration of Tomography Data Acquired before and after Decalcification
2.9. Semi-Automatic Registration of Histology Slides to SRµCT Data
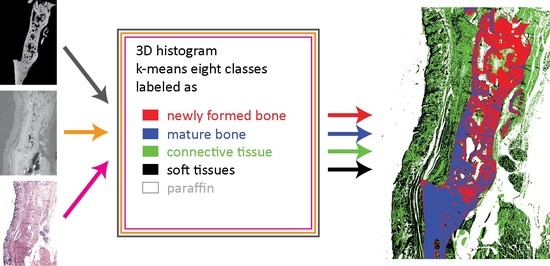
2.10. Joint Histograms, k-Means Clustering, and Segmentation
3. Results and Discussion
3.1. New Bone Formation and the Risk of Non-Union
3.2. Three-Dimensional Imaging of the Distracted Jaws Post Mortem
3.3. Impact of Decalcification on the Hard Tissue Microstructure
3.4. Identifying the Location of the Histology Slide within the Tomography Dataset: 2D-3D Registration
3.5. Univariate Histogram Analysis of the Three Modalities
3.6. Bivariate Histogram Analysis of the Three Modalities
3.7. Trivariate Histogram Analysis of the Three Modalities
3.8. Stem Cell Survival and Differentiation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cope, J.B.; Samchukov, M.L.; Cherkashin, A.M. Mandibular distraction osteogenesis: A historic perspective and future directions. Am. J. Orthod. Dentofac. Orthop. 1999, 115, 448–460. [Google Scholar] [CrossRef]
- McCarthy, J.G.; Stelnicki, E.J.; Mehrara, B.J.; Longaker, M.T. Distraction osteogenesis of the craniofacial skeleton. Plast. Reconstr. Surg. 2001, 107, 1812–1827. [Google Scholar] [CrossRef] [PubMed]
- Heggie, A.A.; Kumar, R.; Shand, J.M. The role of distraction osteogenesis in the management of craniofacial syndromes. Ann. Maxillofac. Surg. 2013, 3, 4–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hariri, F. Distraction Osteogenesis in Oral and Craniomaxillofacial Reconstructive Surgery. In Osteogenesis and Bone Regeneration; IntechOpen: London, UK, 2018. [Google Scholar]
- Felemovicius, J.; Ortiz Monasterio, F.; Gomez Radillo, L.S.; Serna, A. Determining the optimal time for consolidation after distraction osteogenesis. J. Craniomaxillofac. Surg. 2000, 11, 430–436. [Google Scholar] [CrossRef]
- Qi, M.; Hu, J.; Zou, S.; Zhou, H.; Han, L. Mandibular distraction osteogenesis enhanced by bone marrow mesenchymal stem cells in rats. J. Craniomaxillofac. Surg. 2006, 34, 283–289. [Google Scholar] [CrossRef]
- Suehiro, F.; Ishii, M.; Asahina, I.; Murata, H.; Nishimura, M. Low-serum culture with novel medium promotes maxillary/mandibular bone marrow stromal cell proliferation and osteogenic differentiation ability. Clin. Oral Investig. 2017, 21, 2709–2719. [Google Scholar] [CrossRef]
- Ilgenstein, B.; Deyhle, H.; Jaquiery, C.; Kunz, C.; Stalder, A.; Stübinger, S.; Jundt, G.; Beckmann, F.; Müller, B.; Hieber, S.E. Combined micro computed tomography and histology study of bone augmentation and distraction osteogenesis. Proc. SPIE 2012, 8506, 85060M. [Google Scholar] [CrossRef]
- Holme, M.N.; Schulz, G.; Deyhle, H.; Weitkamp, T.; Beckmann, F.; Lobrinus, J.A.; Rikhtegar, F.; Kurtcuoglu, V.; Zanette, I.; Saxer, T.; et al. Complementary X-ray tomography techniques for histology-validated 3D imaging of soft and hard human tissues using plaque-containing blood vessels as examples. Nat. Protoc. 2014, 9, 1401–1415. [Google Scholar] [CrossRef]
- Khimchenko, A.; Schulz, G.; Deyhle, H.; Hieber, S.; Hasan, S.; Bikis, C.; Schulz, G.; Costeur, L.; Müller, B. Non-destructive phase contrast hard X-ray imaging to reveal the three-dimensional microstructure of soft and hard tissues. Proc. SPIE 2016, 9797, 97970B. [Google Scholar] [CrossRef]
- Müller, B.; Lang, S.; Dominietto, M.; Rudin, M.; Schulz, G.; Deyhle, H.; Germann, M.; Pfeiffer, F.; David, C.; Weitkampf, T. High-resolution tomographic imaging of microvessels. Proc. SPIE 2008, 7078, 70780B. [Google Scholar] [CrossRef] [Green Version]
- Müller, B.; Riedel, M.; Thurner, P. Three-dimensional characterization of cell clusters using synchrotron-radiation-based micro computed tomography. Microsc. Microanal. 2006, 12, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campioni, I.; Pecci, R.; Bedini, R. Ten Years of Micro-CT in Dentistry and Maxillofacial Surgery: A Literature Overview. Appl. Sci. 2020, 10, 4328. [Google Scholar] [CrossRef]
- Trejo-Iriarte, C.G.; Serrano-Bello, J.; Gutiérrez-Escalona, R.; Mercado-Marques, C.; García-Honduvilla, N.; Buján-Varela, J.; Medina, L.A. Evaluation of bone regeneration in a critical size cortical bone defect in rat mandible using microCT and histological analysis. Arch. Oral Biol. 2019, 101, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, E.; Paragios, N. Slice-to-volume medical image registration: A survey. Med. Image Anal. 2017, 39, 101–123. [Google Scholar] [CrossRef] [Green Version]
- Stalder, A.; Ilgenstein, B.; Chicherova, N.; Deyhle, H.; Beckmann, F.; Müller, B.; Hieber, S.E. Combined use of micro computed tomography and histology to evaluate the regenerative capacity of bone grafting materials. Int. J. Mater. Res. 2014, 105, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Thurner, P.; Beckmann, F.; Müller, B. An optimization procedure for spatial and density resolution in hard X-ray micro-computed tomography. Nucl. Instrum. Methods Phys. Res. B 2004, 225, 599–603. [Google Scholar] [CrossRef]
- Beckmann, F.; Herzen, J.; Haibel, A.; Müller, B.; Schreyer, A. High density resolution in synchrotron-radiation-based attenuation-contrast microtomography. Proc. SPIE 2008, 7078, 70781D. [Google Scholar] [CrossRef]
- Müller, B.; Bernhardt, R.; Weitkamp, T.; Beckmann, F.; Bräuer, R.; Schurigt, U.; Schrott-Fischer, A.; Glueckert, R.; Ney, M.; Beleites, T.; et al. Morphology of bony tissues and implants uncovered by high-resolution tomographic imaging. Int. J. Mater. Res. 2007, 98, 613–621. [Google Scholar] [CrossRef]
- Müller, B.; Thurner, P.; Beckmann, F.; Weitkamp, T.; Rau, C.; Bernhardt, R.; Karamuk, E.; Eckert, L.; Buchloh, S.; Wintermantel, E.; et al. Nondestructive three-dimensional evaluation of biocompatible materials by microtomography using synchrotron radiation. Proc. SPIE 2002, 4503, 178–188. [Google Scholar] [CrossRef]
- Osinga, R.; Di Maggio, N.; Todorov, A.; Allafi, N.; Barbero, A.; Laurent, F.; Schaefer, D.J.; Martin, I.; Scherberich, A. Generation of a bone organ by human adipose-derived stromal cells through endochondral ossification. Stem Cells Transl. Med. 2016, 5, 1090–1097. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, S.; Staring, M.; Murphy, K.; Viergever, M.A.; Pluim, J.P.W. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 2010, 29, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, S.; Schmidt, M.; Eppler, J.M.; Plesser, H.E.; Masumoto, G.; Igarashi, J.; Ishii, S.; Fukai, T.; Morrison, A.; Diesmann, M.; et al. Spiking network simulation code for petascale computers. Front. Neuroinform. 2014, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Chicherova, N.; Hieber, S.E.; Khimchenko, A.; Bikis, C.; Müller, B.; Cattin, P. Automatic deformable registration of histological slides to μCT volume data. J. Microsc. 2018, 271, 49–61. [Google Scholar] [CrossRef]
- Khimchenko, A.; Deyhle, H.; Schulz, G.; Schweighauser, G.; Hench, J.; Chicherova, N.; Bikis, C.; Hieber, S.; Müller, B. Extending two-dimensional histology into the third dimension through conventional micro computed tomography. Neuroimage 2016, 139, 26–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forgy, E.W. Cluster analysis of multivariate data: Efficiency versus interpretability of classifications. Biometrics 1965, 21, 768–769. [Google Scholar]
- Lloyd, S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Stewart, S.K. Fracture non-union: A review of clinical challenges and future research needs. Malays. Orthop. J. 2019, 13, 1–10. [Google Scholar] [CrossRef]
- Ashman, O.; Phillips, A.M. Treatment of non-unions with bone defects: Which option and why? Injury 2013, 44, S43–S45. [Google Scholar] [CrossRef]
- Toosi, S.; Behravan, N.; Behravan, J. Nonunion fractures, mesenchymal stem cells and bone tissue engineering. J. Biomed. Mater. Res. A. 2018, 106, 2552–2562. [Google Scholar] [CrossRef]
- Becker, K.; Stauber, M.; Schwarz, F.; Beißbarth, T. Automated 3D–2D registration of X-ray microcomputed tomography with histological sections for dental implants in bone using chamfer matching and simulated annealing. Comput. Med. Imaging Graph. 2015, 44, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Lundin, E.L.; Stauber, M.; Papageorgiou, P.; Ehrbar, M.; Ghayor, C.; Weber, F.E.; Tanner, C.; Goksel, O. Automatic registration of 2D histological sections to 3D microCT volumes: Trabecular bone. Bone 2017, 105, 173–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman-de-Sousa, C.; Pinheiro, A.R.; Paramos-de-Carvalho, D.; Costa, M.A.; Ferreirinha, F.; Magalhães-Cardoso, T.; Ribeiro, S.; Pelletier, J.; Sévigny, J.; Correia-de-Sá, P. Opposing effects of adenosine and inosine in human subcutaneous fibroblasts may be regulated by third party ADA cell providers. Cells 2020, 9, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tullberg-Reinert, H.; Jundt, G. In situ measurement of collagen synthesis by human bone cells with a sirius red-based colorimetric microassay: Effects of transforming growth factor beta2 and ascorbic acid 2-phosphate. Histochem. Cell. Biol. 1999, 112, 271–276. [Google Scholar] [CrossRef] [PubMed]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodgers, G.; Sigron, G.R.; Tanner, C.; Hieber, S.E.; Beckmann, F.; Schulz, G.; Scherberich, A.; Jaquiéry, C.; Kunz, C.; Müller, B. Combining High-Resolution Hard X-ray Tomography and Histology for Stem Cell-Mediated Distraction Osteogenesis. Appl. Sci. 2022, 12, 6286. https://doi.org/10.3390/app12126286
Rodgers G, Sigron GR, Tanner C, Hieber SE, Beckmann F, Schulz G, Scherberich A, Jaquiéry C, Kunz C, Müller B. Combining High-Resolution Hard X-ray Tomography and Histology for Stem Cell-Mediated Distraction Osteogenesis. Applied Sciences. 2022; 12(12):6286. https://doi.org/10.3390/app12126286
Chicago/Turabian StyleRodgers, Griffin, Guido R. Sigron, Christine Tanner, Simone E. Hieber, Felix Beckmann, Georg Schulz, Arnaud Scherberich, Claude Jaquiéry, Christoph Kunz, and Bert Müller. 2022. "Combining High-Resolution Hard X-ray Tomography and Histology for Stem Cell-Mediated Distraction Osteogenesis" Applied Sciences 12, no. 12: 6286. https://doi.org/10.3390/app12126286