Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs
Abstract
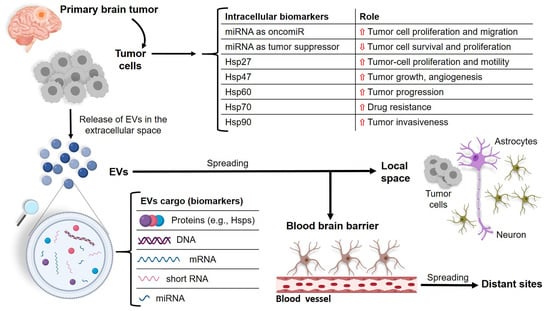
:1. Introduction
2. Intercellular Communication: EVs
3. miRNAs and Molecular Chaperones in Brain Tumors
| Tumor | Cell of Origin | Molecular Features | Clinical Features | Ref. |
|---|---|---|---|---|
| Astrocytomas Diffuse astrocytoma (WHO Grade II); Anaplastic astrocytoma (WHO Grade III); Pilocytic astrocytoma (WHO Grade I); Glioblastoma multiforme (WHO Grade IV); Gliosarcoma (WHO Grade IV) 1. | Astrocyte | p16 deletion p53 mutation PTEN mutation EGFR amplification IDH1 mutation | Astrocytomas, oligodendrogliomas, and ependimomas are gliomas that originate in the glia and are extremely invasive and malignant. Main symptoms are headache, seizures, nausea, vomiting, disturbed vision, tingling sensations, weakness, difficult ambulation. | [52,53,54,55,56,57,58,59,60,61,62,63,64] |
| Oligodendrogliomas Oligodendroglioma (WHO Grade II); -Anaplastic Oligodendroglioma; (WHO Grade III). | Oligodendrocyte | PEG3 deletion EGFR amplification p53 mutation p16 deletion IDH1, IDH2 mutation | ||
| Ependymomas Subependymoma (WHO Grade I); -Myxopapillary ependymoma (WHO Grade I); Ependymoma (WHO Grade II); Ependymoma, RELA fusion-positive (WHO Grade II or III); Anaplastic ependymoma (WHO Grade III). | Ependymal cell | NF2 mutation MT3 underexpression hTERT overexpression miR-485-5p downregulation IGF1 upregulation p16 deletion EGFR amplification | ||
| Meningiomas Meningioma (WHO Grade I); Atypical meningioma (WHO Grade II); Anaplastic meningioma (WHO Grade III). | Meningeal cell | NF2 mutation DAL1 loss PTEN mutation p16 deletion EGFR overexpression | Meningiomas are tumors of the meninges. Main symptoms are headache, seizures, psychotic–motor disabilities, mental weakening, personality changes, visual disorders, language dysfunction. | [37,65,66,67,68,69] |
| Medulloblastomas Medulloblastoma (WHO Grade IV); Desmoplastic/Nodular Medulloblastoma (WHO Grade IV); Medulloblastoma with Extensive Nodularity (WHO Grade IV); Anaplastic Medulloblastoma (WHO Grade IV). | Neuron | p53 mutation TRKC, ERBB2, FSTL5 overexpression PTCH1, CTNNB1 mutation MYC amplification DDX3X mutation | Medulloblastomas are tumors of the cerebellum. Main symptoms are headache, morning vomiting, ataxia. | [70,71,72,73,74,75,76,77,78] |
3.1. miRNAs in Brain Tumors
| Tumor | miRNA | Function | Quantity | Ref. | |
|---|---|---|---|---|---|
| Level | Tissue/Cell | ||||
| Glioma | miR-21 | OncomiR | Increased | GBM cells and derived EVs 1 | [168,169,170,171,172,173,174] |
| Increased | Blood; GBM cells derived EVs | [175] | |||
| miR-148a | OncomiR | Increased | Glioma tissues; glioma cell line; GBM specimens and cell line | [137,138,139,140] | |
| miR-155 | OncomiR | Increased | Blood; Glioma cell line | [141,142,143,144,145] | |
| miR-221/222 | OncomiR | Increased | Blood | [176] | |
| miR-301a | OncomiR | Increased | GBM patient’s serum | [177] | |
| miR-222, miR-124-3p, miR-221, miR-320, miR-574-3p, and miR-301a | Positive diagnostic biomarkers | Increased | Astrocytoma cells | [158] | |
| miR-335 | OncomiR | Increased | GBM cells and derived EVs | [168] | |
| miR-451 | OncomiR | Increased in cells and released in EVs | GBM cells and derived EVs | [178] | |
| miR-1238 | OncomiR | Low levels in cells | GBM cells and derived EVs | [179] | |
| miR-1 | Tumor suppressor | Released in EVs | GBM cells derived EVs | [180] | |
| miR-151a | Tumor suppressor | Low levels | Blood; GBM specimens and cell line | [139,146,149] | |
| miR-185 | Tumor suppressor | Decreased | GBM cell lines and blood | [151,152] | |
| miR-203 | Tumor suppressor | Low levels | Blood | [147,148] | |
| miR-205 | Tumor suppressor | Low levels | GBM tissues and cell lines | [181,182] | |
| miR-454 | Tumor suppressor | Low levels | GMB tissues and cell lines | [153] | |
| miR-605 | Tumor suppressor | Increased | Meningioma tissues | [159] | |
| Meningioma | miR-224 | OncomiR | Increased | Meningioma tissues; meningioma cell | [157] |
| miR-335 | OncomiR | Low levels | Atypical and anaplastic meningiomas; meningioma cell | [156] | |
| miR-145 | Tumor suppressor | Low levels | Benign meningioma tissue | [154,155] | |
| miR-200a | Tumor suppressor | Increased | Malignant meningioma cells | [154,155] | |
| High levels | Blood | [160] | |||
| miR-106a-5p, miR-219-5p, miR-375, and miR-409-3p | Diagnostic and prognostic biomarkers | Low levels | Blood | [160] | |
| miR-197 and miR-224 | Diagnostic and prognostic biomarkers | Increased following recurrence | Tumor samples | [161] | |
| miR-15a-5p, miR-146a-5p, and miR-331-3p | Prognostic biomarkers | [161] | |||
3.2. Molecular Chaperones in Brain Tumors
| Chaperone/Hsp | Role in Gliomas | Ref. |
|---|---|---|
| Hsp27 | Promotes tumor growth, and cancer-cell proliferation and motility | [209,210,211,212,213,214] |
| Hsp47 | Promotes tumor growth, invasiveness, and angiogenesis | [215,216,217] |
| Hsp60 | Promotes tumor progression by enhancing cancer-cell proliferation, preventing apoptosis, and inhibiting the antitumor immune response | [218,219,220] |
| Hsp70 | Promotes cancer cell proliferation, migration, and invasion, and protects cancer cells from apoptosis and anticancer drugs | [227,228,229,230] |
| Hsp90 | Promotes cancer-cell motility, tumor invasiveness, and drug resistance | [231,232,233,234,235] |
| CCT6 | Promotes GBM cell invasion and has a negative association with patient survival 1 | [241] |
4. The Release of miRNAs and Chaperones through EVs as Molecular Signaling and Source of Biomarkers
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, A.; Liu, G. Primary brain tumors in adults: Diagnosis and treatment. Am. Fam. Physician 2016, 93, 211–217. [Google Scholar]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Fucarino, A.; Pitruzzella, A.; David, S.; Campanella, C. Exosomes: Can doctors still ignore their existence? Euromediterranean Biomed. J. 2013, 8, 136–139. [Google Scholar]
- Caruso Bavisotto, C.; Cappello, F.; Macario, A.J.L.; Conway de Macario, E.; Logozzi, M.; Fais, S.; Campanella, C. Exosomal HSP60: A potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev. Mol. Diagn. 2017, 17, 815–822. [Google Scholar] [CrossRef]
- Campanella, C.; Rappa, F.; Sciumè, C.; Marino Gammazza, A.; Barone, R.; Bucchieri, F.; David, S.; Curcurù, G.; Caruso Bavisotto, C.; Pitruzzella, A.; et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 2015, 121, 3230–3239. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Cipolla, C.; Graceffa, G.; Barone, R.; Bucchieri, F.; Bulone, D.; Cabibi, D.; Campanella, C.; Marino Gammazza, A.; Pitruzzella, A.; et al. Immunomorphological pattern of molecular chaperones in normal and pathological thyroid tissues and circulating exosomes: Potential use in clinics. Int. J. Mol. Sci. 2019, 20, 4496. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.U.; Prieto-Vila, M.; Hironaka, A.; Ochiya, T. The role of extracellular vesicle microRNAs in cancer biology. Clin. Chem. Lab. Med. 2017, 55, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.J.; Guerrero-Giménez, M.E.; Prince, T.L.; Ackerman, A.; Bonorino, C.; Calderwood, S.K. Heat shock proteins are essential components in transformation and tumor progression: Cancer cell intrinsic pathways and beyond. Int. J. Mol. Sci. 2019, 20, 4507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graziano, F.; Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C.; Maugeri, R.; Iacopino, D.G. Chaperonology: The third eye on brain gliomas. Brain Sci. 2018, 8, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caruso Bavisotto, C.; Graziano, F.; Rappa, F.; Marino Gammazza, A.; Logozzi, M.; Fais, S.; Maugeri, R.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; et al. Exosomal chaperones and miRNAs in gliomagenesis: State-of-art and theranostics perspectives. Int. J. Mol. Sci. 2018, 19, 2626. [Google Scholar] [CrossRef] [Green Version]
- Marino Gammazza, A.; Caruso Bavisotto, C.; Barone, R.; Conway de Macario, E.; Macario, A.J.L. Alzheimer’s disease and molecular chaperones: Current knowledge and the future of chaperonotherapy. Curr. Pharm. Des. 2016, 22, 4040–4049. [Google Scholar] [CrossRef] [Green Version]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, rna cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes (Basel) 2013, 4, 152–170. [Google Scholar] [CrossRef] [Green Version]
- Perut, F.; Roncuzzi, L.; Baldini, N. The emerging roles of extracellular vesicles in osteosarcoma. Front. Oncol. 2019, 9, 1342. [Google Scholar] [CrossRef] [Green Version]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular vesicle-mediated Cell−Cell communication in the nervous system: Focus on neurological diseases. Int. J. Mol. Sci. 2019, 20, 434. [Google Scholar] [CrossRef] [Green Version]
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular vesicles and their potential use in monitoring cancer progression and therapy: The contribution of proteomics. J. Oncol. 2019, 2019, 1639854. [Google Scholar] [CrossRef]
- Cappello, F.; Logozzi, M.; Campanella, C.; Caruso Bavisotto, C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logozzi, M.; Mizzoni, D.; Bocca, B.; Di Raimo, R.; Petrucci, F.; Caimi, S.; Alimonti, A.; Falchi, M.; Cappello, F.; Campanella, C.; et al. Human primary macrophages scavenge AuNPs and eliminate it through exosomes. A natural shuttling for nanomaterials. Eur. J. Pharm. Biopharm. 2019, 137, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wu, L.-F.; Deng, F.-Y. Exosome: An emerging source of biomarkers for human diseases. Curr. Mol. Med. 2019, 19, 387–394. [Google Scholar] [CrossRef]
- Reiner, A.T.; Witwer, K.W.; Van Balkom, B.W.M.; De Beer, J.; Brodie, C.; Corteling, R.L.; Gabrielsson, S.; Gimona, M.; Ibrahim, A.G.; De Kleijn, D.; et al. Concise review: Developing best-practice models for the therapeutic use of extracellular vesicles. Stem Cells Transl. Med. 2017, 6, 1730–1739. [Google Scholar] [CrossRef] [Green Version]
- Burgio, S.; Noori, L.; Marino Gammazza, A.; Campanella, C.; Logozzi, M.; Fais, S.; Bucchieri, F.; Cappello, F.; Caruso Bavisotto, C. Extracellular vesicles-based drug delivery systems: A new challenge and the exemplum of malignant pleural mesothelioma. Int. J. Mol. Sci. 2020, 21, 5432. [Google Scholar] [CrossRef]
- Nawaz, M.; Malik, M.I.; Hameed, M.; Zhou, J. Research progress on the composition and function of parasite-derived exosomes. Acta Trop. 2019, 196, 30–36. [Google Scholar] [CrossRef]
- Van Dommelen, S.M.; Vader, P.; Lakhal, S.; Kooijmans, S.A.A.; Van Solinge, W.W.; Wood, M.J.A.; Schiffelers, R.M. Microvesicles and exosomes: Opportunities for cell-derived membrane vesicles in drug delivery. J. Control. Release 2012, 161, 635–644. [Google Scholar] [CrossRef]
- Bhome, R.; Del Vecchio, F.; Lee, G.H.; Bullock, M.D.; Primrose, J.N.; Sayan, A.E.; Mirnezami, A.H. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018, 420, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Kleihues, P.; Burger, P.C.; Scheithauer, B.W. The new WHO classification of brain tumours. Brain Pathol. 1993, 3, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Biernat, W. World health organization classification of tumors, of the nervous system. Pol. J. Pathol. 2000, 51, 107–114. [Google Scholar] [PubMed]
- Kleihues, P.; Louis, D.N.; Scheithauer, B.W.; Rorke, L.B.; Reifenberger, G.; Burger, P.C.; Cavenee, W.K. The WHO classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 2002, 61, 215–225. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Posner, J.B.; Chernik, N.L. Intracranial metastases from systemic cancer. Adv. Neurol. 1978, 19, 579–592. [Google Scholar]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Prim. 2019, 5, 5. [Google Scholar] [CrossRef]
- Gaspar, L.E. Brain metastases in lung cancer. Expert Rev. Anticancer 2004, 4, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Gállego Pérez-Larraya, J.; Hildebrand, J. Brain metastases. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 121, pp. 1143–1157. [Google Scholar]
- Michl, M.; Thurmaier, J.; Schubert-Fritschle, G.; Wiedemann, M.; Laubender, R.P.; Nüssler, N.C.; Ruppert, R.; Kleeff, J.; Schepp, W.; Reuter, C.; et al. Brain metastasis in colorectal cancer patients: Survival and analysis of prognostic factors. Clin. Colorectal Cancer 2015, 14, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Witzel, I.; Oliveira-Ferrer, L.; Pantel, K.; Müller, V.; Wikman, H. Breast cancer brain metastases: Biology and new clinical perspectives. Breast Cancer Res. 2016, 18, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemminki, K.; Li, X. Familial risks in nervous system tumors. Cancer Epidemiol. Prev. Biomark. 2003, 12, 1137–1142. [Google Scholar]
- Goh, S.; Butler, W.; Thiele, E.A. Subependymal giant cell tumors in tuberous sclerosis complex. Neurology 2004, 63, 1457–1461. [Google Scholar] [CrossRef]
- Farrell, C.J.; Plotkin, S.R. Genetic causes of brain tumors: Neurofibromatosis, tuberous sclerosis, von hippel-lindau, and other syndromes. Neurol. Clin. 2007, 25, 925–946. [Google Scholar] [CrossRef]
- Hottinger, A.F.; Khakoo, Y. Neuro-oncology of neurofibromatosis type 1. Curr. Treat. Options Neurol. 2009, 11, 306–314. [Google Scholar] [CrossRef]
- Kijima, C.; Miyashita, T.; Suzuki, M.; Oka, H.; Fujii, K. Two cases of nevoid basal cell carcinoma syndrome associated with meningioma caused by a PTCH1 or SUFU germline mutation. Fam. Cancer 2012, 11, 565–570. [Google Scholar] [CrossRef]
- Braganza, M.Z.; Kitahara, C.M.; Berrington De González, A.; Inskip, P.D.; Johnson, K.J.; Rajaraman, P. Ionizing radiation and the risk of brain and central nervous system tumors: A systematic review. Neuro Oncol. 2012, 14, 1316–1324. [Google Scholar] [CrossRef] [Green Version]
- Smoll, N.R.; Brady, Z.; Scurrah, K.; Mathews, J.D. Exposure to ionizing radiation and brain cancer incidence: The life span study cohort. Cancer Epidemiol. 2016, 42, 60–65. [Google Scholar] [CrossRef]
- Libermann, T.A.; Nusbaum, H.R.; Razon, N.; Kris, R.; Lax, I.; Soreq, H.; Whittle, N.; Waterfield, M.D.; Ullrich, A.; Schlessinger, J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature 1985, 313, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Kamb, A.; Gruis, N.A.; Weaver-Feldhaus, J.; Liu, Q.; Harshman, K.; Tavtigian, S.V.; Stockert, E.; Day, R.S.; Johnson, B.E.; Skolnick, M.H. A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264, 436–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, C.; Von Haken, M.; Meyer-Puttlitz, B.; Wiestler, O.D.; Reifenberger, G.; Pietsch, T.; Von Deimling, A. Molecular genetic analysis of ependymal tumors: NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am. J. Pathol. 1999, 155, 627–632. [Google Scholar] [CrossRef]
- Wani, K.; Armstrong, T.S.; Vera-Bolanos, E.; Raghunathan, A.; Ellison, D.; Gilbertson, R.; Vaillant, B.; Goldman, S.; Packer, R.J.; Fouladi, M.; et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012, 123, 727–738. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.; Salunke, P. Understanding Ependymoma oncogenesis: An Update on recent molecular advances and current perspectives. Mol. Neurobiol. 2017, 54, 15–21. [Google Scholar] [CrossRef]
- Li, J.; Yen, C.; Liaw, D.; Podsypanina, K.; Bose, S.; Wang, S.I.; Puc, J.; Miliaresis, C.; Rodgers, L.; McCombie, R.; et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Nigro, J.M.; Baker, S.J.; Preisinger, A.C.; Jessup, J.M.; Hosteller, R.; Cleary, K.; Bigner, S.H.; Davidson, N.; Baylin, S.; Devilee, P.; et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989, 342, 705–708. [Google Scholar] [CrossRef]
- Wolter, M.; Reifenberger, J.; Blaschke, B.; Ichimura, K.; Schmidt, E.E.; Collins, V.P.; Reifenberger, G. Oligodendroglial tumors frequently demonstrate hypermethylation of the CDKN2A (MTS1, p16INK4a), p14ARF, and CDKN2B (MTS2, p15INK4b) tumor suppressor genes. J. Neuropathol. Exp. Neurol. 2001, 60, 1170–1180. [Google Scholar] [CrossRef] [Green Version]
- Bortolotto, S.; Chiadò-Piat, L.; Cavalla, P.; Bosone, I.; Chiò, A.; Mauro, A.; Schiffer, D. CDKN2A/p16 inactivation in the prognosis of oligodendrogliomas. Int. J. Cancer 2000, 88, 554–557. [Google Scholar] [CrossRef]
- Watanabe, T.; Nobusawa, S.; Kleihues, P.; Ohgaki, H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009, 174, 1149–1153. [Google Scholar] [CrossRef] [Green Version]
- Costa, F.F.; Bischof, J.M.; Vanin, E.F.; Lulla, R.R.; Wang, M.; Sredni, S.T.; Rajaram, V.; de Fátima Bonaldo, M.; Wang, D.; Goldman, S.; et al. Identification of micrornas as potential prognostic markers in ependymoma. PloS ONE 2011, 6, e25114. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Poppleton, H.; Fuller, C.; Su, X.; Liu, Y.; Jensen, P.; Magdaleno, S.; Dalton, J.; Calabrese, C.; Board, J.; et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 2005, 8, 323–335. [Google Scholar] [CrossRef] [Green Version]
- Peyre, M.; Commo, F.; Dantas-Barbosa, C.; Andreiuolo, F.; Puget, S.; Lacroix, L.; Drusch, F.; Scott, V.; Varlet, P.; Mauguen, A.; et al. Portrait of ependymoma recurrence in children: Biomarkers of tumor progression identified by dual- color microarray-based gene expression analysis. PloS ONE 2010, 5, e12932. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, C.; Sieberns, J.; Gehlhaar, C.; Simon, M.; Paulus, W.; von Deimling, A. NF2 mutations in secretory and other rare variants of meningiomas. Brain Pathol. 2006, 16, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.H.; Donahoe, J.; Perry, A.; Lemke, N.; Gorse, K.; Kittiniyom, K.; Rempel, S.A.; Gutierrez, J.A.; Newsham, I.F. Loss of DAL-1, a protein 4.1-related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum. Mol. Genet. 2000, 9, 1495–1500. [Google Scholar] [CrossRef] [Green Version]
- Peters, N.; Wellenreuther, R.; Rollbrocker, B.; Hayashi, Y.; Meyer-Puttlitz, B.; Duerr, E.M.; Lenartz, D.; Marsh, D.J.; Schramm, J.; Wiestler, O.D.; et al. Analysis of the PTEN gene in human meningiomas. Neuropathol. Appl. Neurobiol. 1998, 24, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.H.; Smirnov, I.; Baia, G.S.; Modrusan, Z.; Smith, J.S.; Jun, P.; Costello, J.F.; McDermott, M.W.; VandenBerg, S.R.; Lal, A. Molecular signatures define two main classes of meningiomas. Mol. Cancer 2007, 6, 64. [Google Scholar] [CrossRef] [Green Version]
- Arnli, M.B.; Backer-Grøndahl, T.; Ytterhus, B.; Granli, U.S.; Lydersen, S.; Gulati, S.; Torp, S.H. Expression and clinical value of EGFR in human meningiomas. PeerJ 2017, 2017, e3140. [Google Scholar] [CrossRef] [Green Version]
- Pfaff, E.; Remke, M.; Sturm, D.; Benner, A.; Witt, H.; Milde, T.; Von Bueren, A.O.; Wittmann, A.; Schöttler, A.; Jorch, N.; et al. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J. Clin. Oncol. 2010, 28, 5188–5196. [Google Scholar] [CrossRef]
- Hernan, R.; Fasheh, R.; Calabrese, C.; Frank, A.J.; Maclean, K.H.; Allard, D.; Barraclough, R.; Gilbertson, R.J. ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 2003, 63, 140–148. [Google Scholar] [PubMed]
- Grotzer, M.A.; Janss, A.J.; Phillips, P.C.; Trojanowski, J.Q. Neurotrophin receptor TrkC predicts good clinical outcome in medulloblastoma and other primitive neuroectodermal brain tumors. Klin. Padiatr. 2000, 212, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, C.G.; Kratz, J.; Wang, Y.; Summers, K.; Stearns, D.; Cohen, K.; Dang, C.V.; Burger, P.C. Histopathological and molecular prognostic markers in medulloblastoma: C-myc, N-myc, TrkC, and Anaplasia. J. Neuropathol. Exp. Neurol. 2004, 63, 441–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raffel, C.; Jenkins, R.B.; Frederick, L.; Hebrink, D.; Alderete, B.; Fults, D.W.; David James, C. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997, 57, 842–845. [Google Scholar] [PubMed]
- Eberhart, C.G.; Tihan, T.; Burger, P.C. Nuclear localization and mutation of β-catenin in medulloblastomas. J. Neuropathol. Exp. Neurol. 2000, 59, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Gröbner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Northcott, P.A.; Shih, D.J.H.; Peacock, J.; Garzia, L.; Sorana Morrissy, A.; Zichner, T.; Stútz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012, 487, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Tantravedi, S.; Vesuna, F.; Winnard, P.T.; Martin, A.; Lim, M.; Eberhart, C.G.; Berlinicke, C.; Raabe, E.; van Diest, P.J.; Raman, V. Targeting DDX3 in medulloblastoma using the small molecule inhibitor RK-33. Transl. Oncol. 2019, 12, 96–105. [Google Scholar] [CrossRef]
- Sanai, N.; Polley, M.Y.; Berger, M.S. Insular glioma resection: Assessment of patient morbidity, survival, and tumor progression—Clinical article. J. Neurosurg. 2010, 112, 1–9. [Google Scholar] [CrossRef]
- Ius, T.; Isola, M.; Budai, R.; Pauletto, G.; Tomasino, B.; Fadiga, L.; Skrap, M. Low-grade glioma surgery in eloquent areas: Volumetric analysis of extent of resection and its impact on overall survival. A single-institution experience in 190 patients—Clinical article. J. Neurosurg. 2012, 117, 1039–1052. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21, V1–V100. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Fisher, J.L.; Nichols, E.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; Abraha, H.N.; Agius, D.; Alahdab, F.; Alam, T.; et al. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 376–393. [Google Scholar] [CrossRef] [Green Version]
- Osei, E.; Walters, P.; Masella, O.; Tennant, Q.; Fishwick, A.; Dadzie, E.; Bhangu, A.; Darko, J. A review of predictive, prognostic and diagnostic biomarkers for brain tumours: Towards personalised and targeted cancer therapy. J. Radiother. Pract. 2019, 1–16. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketting, R.F.; Fischer, S.E.J.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H.A. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef]
- Olsen, P.H.; Ambros, V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999, 216, 671–680. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Seggerson, K.; Tang, L.; Moss, E.G. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002, 243, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- miRbase. Available online: http://www.mirbase.org/help/nomenclature.shtml (accessed on 23 September 2020).
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Grad, Y.; Aach, J.; Hayes, G.D.; Reinhart, B.J.; Church, G.M.; Ruvkun, G.; Kim, J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell 2003, 11, 1253–1263. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. The microRNA registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Huang, T.H.; Fan, B.; Rothschild, M.F.; Hu, Z.L.; Li, K.; Zhao, S.H. MiRFinder: An improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinform. 2007, 8, 341. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Sethupathy, P.; Megraw, M.; Hatzigeorgiou, A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 2006, 3, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, W. MicroRNA target prediction. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1513, pp. 193–200. [Google Scholar]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Sugatani, T.; Hruska, K.A. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 2009, 284, 4667–4678. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Jin, Y.X. MicroRNA in cell differentiation and development. Sci. China Ser. C Life Sci. 2009, 52, 205–211. [Google Scholar] [CrossRef]
- Tzur, G.; Israel, A.; Levy, A.; Benjamin, H.; Meiri, E.; Shufaro, Y.; Meir, K.; Khvalevsky, E.; Spector, Y.; Rojansky, N.; et al. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PloS ONE 2009, 4, e7511. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic MicroRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Wong, Q.W.L.; Lung, R.W.M.; Law, P.T.Y.; Lai, P.B.S.; Chan, K.Y.Y.; To, K.F.; Wong, N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of stathmin1. Gastroenterology 2008, 135, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U.; Hasemeier, B.; Christgen, M.; Müller, M.; Römermann, D.; Länger, F.; Kreipe, H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008, 214, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walz, A.L.; Ooms, A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Guidry Auvil, J.M.; Meerzaman, D.; Chen, Q.-R.; Hsu, C.H.; Yan, C.; et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliou, M.S.; Da Silva-Diz, V.; Carmona, F.J.; Ramalho-Carvalho, J.; Heyn, H.; Villanueva, A.; Muñoz, P.; Esteller, M. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene 2014, 33, 4003–4015. [Google Scholar] [CrossRef] [Green Version]
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef]
- Shi, L.; Cheng, Z.; Zhang, J.; Li, R.; Zhao, P.; Fu, Z.; You, Y. Hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008, 1236, 185–193. [Google Scholar] [CrossRef]
- Li, W.B.; Ma, M.W.; Dong, L.J.; Wang, F.; Chen, L.X.; Li, X.R. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol. 2011, 12, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xiong, Y.; Zhao, Y.; Tao, L.; Zhang, Z.; Zhang, H.; Liu, Y.; Feng, G.; Li, B.; He, L.; et al. Identification of aberrant microRNA expression pattern in pediatric gliomas by microarray. Diagn. Pathol. 2013, 8, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.P.; Mou, K.J.; Xu, Q.F.; Tang, J.H.; Huang, G.H.; Xu, J.P.; Li, G.H.; Ai, S.J.; Hugnot, J.P.; Zhou, Z.; et al. Microarray analysis of the aberrant microRNA expression pattern in gliomas of different grades. Oncol. Rep. 2015, 34, 318–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Shi, H.; Lai, N.; Liao, K.; Zhang, S.; Lu, X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med. Oncol. 2014, 31, 911. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zou, R.; Zhou, R.; Gong, C.; Wang, Z.; Cai, T.; Tan, C.; Fang, J. MiR-155 regulates glioma cells invasion and chemosensitivity by p38 isforms in vitro. J. Cell. Biochem. 2015, 116, 1213–1221. [Google Scholar] [CrossRef]
- Wu, W.; Yu, T.; Wu, Y.; Tian, W.; Zhang, J.; Wang, Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J. Exp. Clin. Cancer Res. 2019, 38, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Chen, Z.; Zhao, H. MicroRNA-155-3p promotes glioma progression and temozolomide resistance by targeting Six1. J. Cell. Mol. Med. 2020, 24, 5363–5374. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, J.; Hao, J.; Shi, Z.; Wang, Y.; Han, L.; Yu, S.; You, Y.; Jiang, T.; Wang, J.; et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 Family as novel descriptive and prognostic biomarkers for glioma. Mol. Neurobiol. 2016, 53, 1452–1460. [Google Scholar] [CrossRef]
- Zhang, J.; Han, L.; Ge, Y.; Zhou, X.; Zhang, A.; Zhang, C.; Zhong, Y.; You, Y.; Pu, P.; Kang, C. MiR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int. J. Oncol. 2010, 36, 913–920. [Google Scholar]
- Zhang, C.; Kang, C.; You, Y.; Pu, P.; Yang, W.; Zhao, P.; Wang, G.; Wang, A.; Jia, Z.; Han, L.; et al. Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int. J. Oncol. 2009, 34, 1653–1660. [Google Scholar] [PubMed] [Green Version]
- Chen, L.; Zhang, J.; Han, L.; Zhang, A.; Zhang, C.; Zheng, Y.; Jiang, T.; Pu, P.; Jiang, C.; Kang, C. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol. Rep. 2012, 27, 854–860. [Google Scholar] [PubMed]
- Tang, H.; Liu, Q.; Liu, X.; Ye, F.; Xie, X.; Xie, X.; Wu, M. Plasma miR-185 as a predictive biomarker for prognosis of malignant glioma. J. Cancer Res. 2015, 11, 630–634. [Google Scholar]
- Yue, X.; Lan, F.; Hu, M.; Pan, Q.; Wang, Q.; Wang, J. Downregulation of serum microRNA-205 as a potential diagnostic and prognostic biomarker for human glioma. J. Neurosurg. 2016, 124, 122–128. [Google Scholar] [CrossRef]
- Yue, X.; Wang, P.; Xu, J.; Zhu, Y.; Sun, G.; Pang, Q.; Tao, R. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol. Rep. 2012, 27, 1200–1206. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Wang, Z.; Liu, X.; Liu, Q.; Xu, G.; Li, G.; Wu, M. LRRC4 inhibits glioma cell growth and invasion through a miR-185- dependent pathway. Curr. Cancer Drug Targets 2012, 12, 1032–1042. [Google Scholar] [CrossRef]
- Batash, R.; Asna, N.; Schaffer, P.; Francis, N.; Schaffer, M. Glioblastoma multiforme, diagnosis and treatment; Recent literature review. Curr. Med. Chem. 2017, 24, 3002–3009. [Google Scholar] [CrossRef]
- Chen, J.; Yang, L.; Wang, X. Reduced circulating microRNA-203 predicts poor prognosis for glioblastoma. Cancer Biomark. 2017, 20, 521–526. [Google Scholar] [CrossRef]
- Liao, H.; Bai, Y.; Qiu, S.; Zheng, L.; Huang, L.; Liu, T.; Wang, X.; Liu, Y.; Xu, N.; Yan, X.; et al. MiR-203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial-mesenchymal transition via SNAI2. Oncotarget 2015, 6, 8914–8928. [Google Scholar] [CrossRef]
- Jia, J.; Wang, J.; Yin, M.; Liu, Y. MicroRNA-605 directly targets sox9 to alleviate the aggressive phenotypes of glioblastoma multiforme cell lines by deactivating the PI3K/Akt pathway. Onco Targets Ther. 2019, 12, 5437–5448. [Google Scholar] [CrossRef] [Green Version]
- Saydam, O.; Shen, Y.; Würdinger, T.; Senol, O.; Boke, E.; James, M.F.; Tannous, B.A.; Stemmer-Rachamimov, A.O.; Yi, M.; Stephens, R.M.; et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway. Mol. Cell. Biol. 2009, 29, 5923–5940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senol, O.; Schaaij-Visser, T.B.M.; Erkan, E.P.; Dorfer, C.; Lewandrowski, G.; Pham, T.V.; Piersma, S.R.; Peerdeman, S.M.; Ströbel, T.; Tannous, B.; et al. MiR-200a-mediated suppression of non-muscle heavy chain IIb inhibits meningioma cell migration and tumor growth in vivo. Oncogene 2014, 34, 1790–1798. [Google Scholar] [CrossRef] [PubMed]
- Kliese, N.; Gobrecht, P.; Pachow, D.; Andrae, N.; Wilisch-Neumann, A.; Kirches, E.; Riek-Burchardt, M.; Angenstein, F.; Reifenberger, G.; Riemenschneider, M.; et al. MiRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene 2013, 32, 4712–4720. [Google Scholar] [CrossRef]
- Shi, L.; Jiang, D.; Sun, G.; Wan, Y.; Zhang, S.; Zeng, Y.; Pan, T.; Wang, Z. MiR-335 promotes cell proliferation by directly targeting Rb1 in meningiomas. J. Neurooncol. 2012, 110, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Zheng, X.; Wu, S.; Lu, H.; Leng, T.; Zhu, W.; Zhou, Y.; Ou, Y.; Lin, X.; Lin, Y.; et al. Targeting oncogenic miR-335 inhibits growth and invasion of malignant astrocytoma cells. Mol. Cancer 2011, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Deng, X.; Ying, Q.; Jin, T.; Li, M.; Liang, C. MicroRNA-224 targets ERG2 and contributes to malignant progressions of meningioma. Biochem. Biophys. Res. Commun. 2015, 460, 354–361. [Google Scholar] [CrossRef]
- Zhi, F.; Shao, N.; Li, B.; Xue, L.; Deng, D.; Xu, Y.; Lan, Q.; Peng, Y.; Yang, Y. A serum 6-miRNA panel as a novel non-invasive biomarker for meningioma. Sci. Rep. 2016, 6, 32067. [Google Scholar] [CrossRef] [Green Version]
- Slavik, H.; Balik, V.; Vrbkova, J.; Rehulkova, A.; Vaverka, M.; Hrabalek, L.; Ehrmann, J.; Vidlarova, M.; Gurska, S.; Hajduch, M.; et al. Identification of meningioma patients at high risk of tumor recurrence using microRNA profiling. Neurosurgery 2020, 3, nyaa009. [Google Scholar] [CrossRef]
- Xiao, F.; Lv, S.; Zong, Z.; Wu, L.; Tang, X.; Kuang, W.; Zhang, P.; Li, X.; Fu, J.; Xiao, M.; et al. Cerebrospinal fluid biomarkers for brain tumor detection: Clinical roles and current progress. Am. J. Transl. Res. 2020, 12, 1379–1396. [Google Scholar]
- Drusco, A.; Bottoni, A.; Lagana’, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M.P.; et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef] [Green Version]
- Petrescu, G.E.D.; Sabo, A.A.; Torsin, L.I.; Calin, G.A.; Dragomir, M.P. MicroRNA based theranostics for brain cancer: Basic principles. J. Exp. Clin. Cancer Res. 2019, 38, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zöllner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012, 14, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kopkova, A.; Sana, J.; Machackova, T.; Vecera, M.; Radova, L.; Trachtova, K.; Vybihal, V.; Smrcka, M.; Kazda, T.; Slaby, O.; et al. Cerebrospinal fluid microRNA signatures as diagnostic biomarkers in brain tumors. Cancers 2019, 11, 1546. [Google Scholar] [CrossRef] [Green Version]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012, 14, 689–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.W.; Skog, J.; et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. 2016, 18, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abels, E.R.; Maas, S.L.N.; Nieland, L.; Wei, Z.; Cheah, P.S.; Tai, E.; Kolsteeg, C.J.; Dusoswa, S.A.; Ting, D.T.; Hickman, S.; et al. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 2019, 28, 3105–3119.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 targets a network of key tumor-suppressive Pathways in glioblastoma cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Ma, X.; Wang, J.; Zhao, Y.; Wang, Y.; Bihl, J.C.; Chen, Y.; Jiang, C. Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21/VEGF signal. Oncotarget 2017, 8, 36137–36148. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): A platform for glioblastoma biomarker development. PloS ONE 2013, 8, e78115. [Google Scholar] [CrossRef]
- Shi, R.; Wang, P.Y.; Li, X.Y.; Chen, J.X.; Li, Y.; Zhang, X.Z.; Zhang, C.G.; Jiang, T.; Li, W.B.; Ding, W.; et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Zhu, A.; Gong, L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bull. Cancer 2018, 105, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Qing, Q.; Pan, Q.; Hu, M.; Yu, H.; Yue, X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell. Oncol. 2018, 41, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Manterola, L.; Guruceaga, E.; Gállego Pérez-Larraya, J.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zeng, A.; Zhang, Z.; Shi, Z.; Yan, W.; You, Y. Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine 2019, 42, 238–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronisz, A.; Wang, Y.; Nowicki, M.O.; Peruzzi, P.; Ansari, K.I.; Ogawa, D.; Balaj, L.; De Rienzo, G.; Mineo, M.; Nakano, I.; et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014, 74, 738–750. [Google Scholar] [CrossRef] [Green Version]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef]
- Fang, B.; Zhu, J.; Wang, Y.; Geng, F.; Li, G. MiR-454 inhibited cell proliferation of human glioblastoma cells by suppressing PDK1 expression. Biomed. Pharm. 2015, 75, 148–152. [Google Scholar] [CrossRef]
- Shao, N.; Xue, L.; Wang, R.; Luo, K.; Zhi, F.; Lan, Q. MiR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol. Cancer 2019, 18, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Macario, A.J.L.; Conway de Macario, E. Chaperone proteins and chaperonopathies. In Stress: Physiology, Biochemistry, and Pathology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 135–152. [Google Scholar]
- Dahiya, V.; Buchner, J. Functional principles and regulation of molecular chaperones. In Advances in Protein Chemistry and Structural Biology; Academic Press Inc.: Cambridge, MA, USA, 2019; Volume 114, pp. 1–60. ISBN 9780128155578. [Google Scholar]
- Hendrick, J.P.; Hartl, F.U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef]
- Becker, J.; Craig, E.A. Heat-shock proteins as molecular chaperones. Eur. J. Biochem. 1994, 219, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Nollen, E.A.; Morimoto, R.I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing “heat shock” proteins. J. Cell Sci. 2002, 115, 2809–2816. [Google Scholar] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant. Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. Proteins as molecular chaperones. Nature 1988, 328, 378–379. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. The molecular chaperone concept. Semin. Cell Biol. 1990, 1, 1–9. [Google Scholar]
- Ellis, R.J. Molecular chaperones: Assisting assembly in addition to folding. Trends Biochem. Sci. 2006, 31, 395–401. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Finka, A.; Sharma, S.K.; Goloubinoff, P. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front. Mol. Biosci. 2015, 2, 29. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef]
- Willison, K.R. The structure and evolution of eukaryotic chaperonin-containing TCP-1 and its mechanism that folds actin into a protein spring. Biochem. J. 2018, 475, 3009–3034. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.M.; Oster, M.E.; Hebert, D.N. Protein quality control in the endoplasmic reticulum. Protein J. 2019, 38, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Juste, Y.R.; Cuervo, A.M. Analysis of chaperone-mediated autophagy. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 1880, pp. 703–727. [Google Scholar]
- Pockley, A.G.; Henderson, B. Extracellular cell stress (Heat shock) proteins—immune responses and disease: An overview. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, B.; Calderwood, S.K.; Coates, A.R.M.; Cohen, I.; van Eden, W.; Lehner, T.; Pockley, A.G. Caught with their PAMPs down? The extracellular signalling actions of molecular chaperones are not due to microbial contaminants. Cell Stress Chaperones 2010, 15, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Henderson, B.; Pockley, A.G. Molecular chaperones and protein-folding catalysts as intercellular signaling regulators in immunity and inflammation. J. Leukoc. Biol. 2010, 88, 445–462. [Google Scholar] [CrossRef]
- Macario, A.J.L.; Conway de Macario, E. Sick chaperones, cellular stress, and disease. New Engl. J. Med. 2005, 353, 1489–1501. [Google Scholar] [CrossRef]
- Macario, A.J.L.; Conway de Macario, E.; Cappello, F. The Chaperonopathies. Diseases with Defective Molecular Chaperones; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Hermisson, M.; Strik, H.; Rieger, J.; Dichgans, J.; Meyermann, R.; Weller, M. Expression and functional activity of heat shock proteins in human glioblastoma multiforme. Neurology 2000, 54, 1357–1364. [Google Scholar] [CrossRef]
- Yang, I.; Fang, S.; Parsa, A.T. Heat shock proteins in glioblastomas. Neurosurg. Clin. 2010, 21, 111–123. [Google Scholar] [CrossRef]
- Rajesh, Y.; Biswas, A.; Mandal, M. Glioma progression through the prism of heat shock protein mediated extracellular matrix remodeling and epithelial to mesenchymal transition. Exp. Cell Res. 2017, 359, 299–311. [Google Scholar] [CrossRef]
- Iglesia, R.P.; de Lima Fernandes, C.F.; Coelho, B.P.; Prado, M.B.; Escobar, M.I.M.; Almeida, G.H.D.R.; Lopes, M.H. Heat shock proteins in glioblastoma biology: Where do we stand? Int. J. Mol. Sci. 2019, 20, 5794. [Google Scholar] [CrossRef] [Green Version]
- Alexiou, G.A. Role of heat shock proteins in brain tumors. In Heat Shock Proteins in Neuroscience; Springer International Publishing: Cham, Switzerland, 2019; pp. 23–28. [Google Scholar]
- Khalid, H.; Tsutsumi, K.; Yamashita, H.; Kishikawa, M.; Yasunaga, A.; Shibata, S. Expression of the small heat shock protein (hsp) 27 in human astrocytomas correlates with histologic grades and tumor growth fractions. Cell. Mol. Neurobiol. 1995, 15, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Assimakopoulou, M.; Sotiropoulou-Bonikou, G.; Maraziotis, T.; Varakis, I. Prognostic significance of Hsp-27 in astrocytic brain tumors: An immunohistochemical study. Anticancer Res. 1997, 17, 2677–2682. [Google Scholar] [PubMed]
- Nomura, N.; Nomura, M.; Sugiyama, K.; Hamada, J.I. Phorbol 12-myristate 13-acetate (PMA)-induced migration of glioblastoma cells is mediated via p38MAPK/Hsp27 pathway. Biochem. Pharm. 2007, 74, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Sang, D.P.; Li, R.J.; Lan, Q. Quercetin sensitizes human glioblastoma cells to temozolomide in vitro via inhibition of Hsp27. Acta Pharm. Sin. 2014, 35, 832–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belkacemi, L.; Hebb, M.O. HSP27 knockdown produces synergistic induction of apoptosis by HSP90 and kinase inhibitors in glioblastoma multiforme. Anticancer Res. 2014, 34, 4915–4927. [Google Scholar] [PubMed]
- Önay Uçar, E.; Şengelen, A. Resveratrol and siRNA in combination reduces Hsp27 expression and induces caspase-3 activity in human glioblastoma cells. Cell Stress Chaperones 2019, 24, 763–775. [Google Scholar] [CrossRef]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Leng, Z.G.; Guo, Y.H.; Yang, W.L.; Chu, Y.W.; Yang, S.-H.; Zhao, W.G. Heat shock protein 47 promotes glioma angiogenesis. Brain Pathol. 2016, 26, 31–42. [Google Scholar] [CrossRef]
- Zhao, D.; Jiang, X.; Yao, C.; Zhang, L.; Liu, H.; Xia, H.; Wang, Y. Heat shock protein 47 regulated by miR-29a to enhance glioma tumor growth and invasion. J. Neurooncol. 2014, 118, 39–47. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, T.; Wang, Z.; Qi, B.; Xia, H. HSP47 promotes glioblastoma stemlike cell survival by modulating tumor microenvironment extracellular matrix through TGF-β pathway. Acs Chem. Neurosci. 2017, 8, 128–134. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Siegelin, M.D.; Dohi, T.; Altieri, D.C. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010, 70, 8988–8993. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Li, J.; Liu, X.; Wang, G.; Luo, M.; Deng, H. Down-regulation of HSP60 suppresses the proliferation of glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci. Rep. 2016, 6, 28388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappa, F.; Unti, E.; Baiamonte, P.; Cappello, F.; Scibetta, N. Different immunohistochemical levels of Hsp60 and Hsp70 in a subset of brain tumors and putative role of Hsp60 in neuroepithelial tumorigenesis. Eur. J. Histochem. 2013, 57, e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Cascio, F.L.; Mocciaro, E.; Vitale, A.M.; Vergilio, G.; Pace, A.; Cappello, F.; Campanella, C.; Palumbo Piccionello, A. Curcumin affects HSP60 folding activity and levels in neuroblastoma cells. Int. J. Mol. Sci. 2020, 21, 661. [Google Scholar] [CrossRef] [Green Version]
- Marino Gammazza, A.; Caruso Bavisotto, C.; David, S.; Barone, R.; Rappa, F.; Campanella, C.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L. HSP60 is a ubiquitous player in the physiological and pathogenic interactions between the chaperoning and the immune systems. Curr. Immunol. Rev. 2017, 13, 44–55. [Google Scholar]
- Caruso Bavisotto, C.; Alberti, G.; Vitale, A.M.; Paladino, L.; Campanella, C.; Rappa, F.; Gorska, M.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L.; et al. Hsp60 Post-translational Modifications: Functional and Pathological Consequences. Front. Mol. Biosci. 2020, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Vilasi, S.; Bulone, D.; Caruso Bavisotto, C.; Campanella, C.; Marino Gammazza, A.; San Biagio, P.L.; Cappello, F.; Conway de Macario, E.; Macario, A.J.L. Chaperonin of group I: Oligomeric spectrum and biochemical and biological implications. Front. Mol. Biosci. 2018, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Albakova, Z.; Armeev, G.A.; Kanevskiy, L.M.; Kovalenko, E.I.; Sapozhnikov, A.M. HSP70 multi-functionality in cancer. Cells 2020, 9, 587. [Google Scholar] [CrossRef] [Green Version]
- Thorsteinsdottir, J.; Stangl, S.; Fu, P.; Guo, K.; Albrecht, V.; Eigenbrod, S.; Erl, J.; Gehrmann, M.; Tonn, J.C.; Multhoff, G.; et al. Overexpression of cytosolic, plasma membrane bound and extracellular heat shock protein 70 (Hsp70) in primary glioblastomas. J. Neurooncol. 2017, 135, 443–452. [Google Scholar] [CrossRef]
- Da Rocha, A.B.; Regner, A.; Grivicich, I.; Pretto Schunemann, D.; Diel, C.; Kovaleski, G.; De Farias, C.B.; Mondadori, E.; Almeida, L.; Braga Filho, A.; et al. Radioresistance is associated to increased Hsp70 content in human glioblastoma cell lines. Int. J. Oncol. 2004, 25, 777–785. [Google Scholar] [CrossRef]
- Sun, G.; Cao, Y.; Xu, Y.; Huai, D.; Chen, P.; Guo, J.; Li, M.; Dai, Y. Overexpression of Hsc70 promotes proliferation, migration, and invasion of human glioma cells. J. Cell. Biochem. 2019, 120, 10707–10714. [Google Scholar] [CrossRef]
- Li, G.; Xu, Y.; Guan, D.; Liu, Z.; Liu, D.X. HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J. Biol. Chem. 2011, 286, 20251–20259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarev, V.F.; Nikotina, A.D.; Mikhaylova, E.R.; Nudler, E.; Polonik, S.G.; Guzhova, I.V.; Margulis, B.A. Hsp70 chaperone rescues C6 rat glioblastoma cells from oxidative stress by sequestration of aggregating GAPDH. Biochem. Biophys. Res. Commun. 2016, 470, 766–771. [Google Scholar] [CrossRef]
- Gopal, U.; Bohonowych, J.E.; Lema-Tome, C.; Liu, A.; Garrett-Mayer, E.; Wang, B.; Isaacs, J.S. A novel extracellular Hsp90 mediated co-receptor function for LRP1 regulates EphA2 dependent glioblastoma cell invasion. PloS ONE 2011, 6, e17649. [Google Scholar] [CrossRef] [PubMed]
- Thuringer, D.; Hammann, A.; Benikhlef, N.; Fourmaux, E.; Bouchot, A.; Wettstein, G.; Solary, E.; Garrido, C. Transactivation of the epidermal growth factor receptor by heat shock protein 90 via toll-like receptor 4 contributes to the migration of glioblastoma cells. J. Biol. Chem. 2011, 286, 3418–3428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memmel, S.; Sisario, D.; Zöller, C.; Fiedler, V.; Katzer, A.; Heiden, R.; Becker, N.; Eing, L.; Ferreira, F.L.R.; Zimmermann, H.; et al. Migration pattern, actin cytoskeleton organization and response to PI3K-, mTOR-, and Hsp90-inhibition of glioblastoma cells with different invasive capacities. Oncotarget 2017, 8, 45298–45310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snigireva, A.V.; Vrublevskaya, V.V.; Skarga, Y.Y.; Morenkov, O.S. Cell surface heparan sulfate proteoglycans are involved in the extracellular Hsp90-stimulated migration and invasion of cancer cells. Cell Stress Chaperones 2019, 24, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Wachsberger, P.R.; Lawrence, Y.R.; Liu, Y.; Rice, B.; Feo, N.; Leiby, B.; Dicker, A.P. Hsp90 inhibition enhances PI-3 kinase inhibition and radiosensitivity in glioblastoma. J. Cancer Res. Clin. Oncol. 2014, 140, 573–582. [Google Scholar] [CrossRef]
- Sauvageot, C.M.-E.; Weatherbee, J.L.; Kesari, S.; Winters, S.E.; Barnes, J.; Dellagatta, J.; Ramakrishna, N.R.; Stiles, C.D.; Kung, A.L.-J.; Kieran, M.W.; et al. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol. 2009, 11, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Woolfenden, S.; Bronson, R.T.; Jaffer, Z.M.; Barluenga, S.; Winssinger, N.; Rubenstein, A.E.; Chen, R.; Charest, A. The novel Hsp90 inhibitor NXD30001 induces tumor regression in a genetically engineered mouse model of glioblastoma multiforme. Mol. Cancer Ther. 2010, 9, 2618–2626. [Google Scholar] [CrossRef] [Green Version]
- Canella, A.; Welker, A.M.; Yoo, J.Y.; Xu, J.; Abas, F.S.; Kesanakurti, D.; Nagarajan, P.; Beattie, C.E.; Sulman, E.P.; Liu, J.; et al. Efficacy of onalespib, a long-acting second-generation HSP90 inhibitor, as a single agent and in combination with temozolomide against malignant gliomas. Clin. Cancer Res. 2017, 23, 6215–6226. [Google Scholar] [CrossRef] [Green Version]
- Hauser, P.; Hanzély, Z.; Jakab, Z.; Oláh, L.; Szabó, E.; Jeney, A.; Schuler, D.; Fekete, G.; Bognár, L.; Garami, M. Expression and prognostic examination of heat shock proteins (HSP 27, HSP 70, and HSP 90) in medulloblastoma. J. Pediatr. Hematol. Oncol. 2006, 28, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, G.A.; Vartholomatos, G.; Stefanaki, K.; Patereli, A.; Dova, L.; Karamoutsios, A.; Lallas, G.; Sfakianos, G.; Moschovi, M.; Prodromou, N. Expression of heat shock proteins in medulloblastoma: Laboratory investigation. J. Neurosurg. Pediatr. 2013, 12, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Russell, B.P.; Wei, H.; Lee, M.Y.T.; Toon, C.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Kaufman, K.L. Extracellular vesicles from neurosurgical aspirates identifies chaperonin containing TCP1 subunit 6A as a potential glioblastoma biomarker with prognostic significance. Proteomics 2019, 19, e1800157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graner, M.W.; Cumming, R.I.; Bigner, D.D. The heat shock response and chaperones/heat shock proteins in brain tumors: Surface expression, release, and possible immune consequences. J. Neurosci. 2007, 27, 11214–11227. [Google Scholar] [CrossRef]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23, 1541–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epple, L.M.; Griffiths, S.G.; Dechkovskaia, A.M.; Dusto, N.L.; White, J.; Ouellette, R.J.; Anchordoquy, T.J.; Bemis, L.T.; Graner, M.W. Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS ONE 2012, 7, e42064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graner, M.; Redzic, J.; Ung, T. Glioblastoma extracellular vesicles: Reservoirs of potential biomarkers. Pharmgenomics Pers. Med. 2014, 7, 65. [Google Scholar] [CrossRef] [Green Version]
- Lane, R.; Simon, T.; Vintu, M.; Solkin, B.; Koch, B.; Stewart, N.; Benstead-Hume, G.; Pearl, F.M.G.; Critchley, G.; Stebbing, J.; et al. Cell-derived extracellular vesicles can be used as a biomarker reservoir for glioblastoma tumor subtyping. Commun. Biol. 2019, 2, 315. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Bronisz, A.; Godlewski, J.; Chiocca, E.A. Extracellular vesicles and microRNAs: Their role in Tumorigenicity and therapy for brain tumors. Cell. Mol. Neurobiol. 2016, 36, 361–376. [Google Scholar] [CrossRef]
- Gourlay, J.; Morokoff, A.P.; Luwor, R.B.; Zhu, H.J.; Kaye, A.H.; Stylli, S.S. The emergent role of exosomes in glioma. J. Clin. Neurosci. 2017, 35, 13–23. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Z.; Mashimo, T.; Shen, B.; Nyagilo, J.; Wang, H.; Wang, Y.; Liu, Z.; Mulgaonkar, A.; Hu, X.L.; et al. Gliomas interact with non-glioma brain cells via extracellular vesicles. Cell Rep. 2020, 30, 2489–2500.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, A.L.; Yan, W.; Liu, Y.W.; Wang, Z.; Hu, Q.; Nie, E.; Zhou, X.; Li, R.; Wang, X.F.; Jiang, T.; et al. Tumour exosomes from cells harbouring PTPRZ1-MET fusion contribute to a malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene 2017, 36, 5369–5381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lötvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Zhu, H.; Zhang, M.; Xu, J. Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5. Int. J. Mol. Med. 2018, 41, 155–163. [Google Scholar] [CrossRef]
- Tytell, M. Release of heat shock proteins (Hsps) and the effects of extracellular Hsps on neural cells and tissues. Int. J. Hyperth. 2005, 21, 445–455. [Google Scholar] [CrossRef]
- Van Den Boorn, J.G.; Schlee, M.; Coch, C.; Hartmann, G. SiRNA delivery with exosome nanoparticles. Nat. Biotechnol. 2011, 29, 325–326. [Google Scholar] [CrossRef]
- Pullan, J.E.; Confeld, M.I.; Osborn, J.K.; Kim, J.; Sarkar, K.; Mallik, S. Exosomes as drug carriers for cancer therapy. Mol. Pharm. 2019, 16, 1789–1798. [Google Scholar] [CrossRef]
- Kim, G.; Kim, M.; Lee, Y.; Byun, J.W.; Hwang, D.W.; Lee, M. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J. Control. Release 2020, 317, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Bu, N.; Wu, H.; Sun, B.; Zhang, G.; Zhan, S.; Zhang, R.; Zhou, L. Exosome-loaded dendritic cells elicit tumor-specific CD8 + cytotoxic T cells in patients with glioma. J. Neurooncol. 2011, 104, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Bu, N.; Wu, H.; Zhang, G.; Zhan, S.; Zhang, R.; Sun, H.; Du, Y.; Yao, L.; Wang, H. Exosomes from dendritic cells loaded with chaperone-rich cell lysates elicit a potent T cell immune response Against intracranial glioma in mice. J. Mol. Neurosci. 2015, 56, 631–643. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.F.; O’Driscoll, L. MiR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.F.; Chang, Y.W.; Andreu-Vieyra, C.; Fang, J.Y.; Yang, Z.; Han, B.; Lee, A.S.; Liang, G. MiR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene 2013, 32, 4694–4701. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Xue, Y.; Wang, P.; Wang, Z.; Li, Z.; Hu, Y.; Li, Z.; Shang, X.; Liu, Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 2015, 6, 19759–19779. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, A.M.; Santonocito, R.; Vergilio, G.; Marino Gammazza, A.; Campanella, C.; Conway de Macario, E.; Bucchieri, F.; Macario, A.J.L.; Caruso Bavisotto, C. Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs. Appl. Sci. 2020, 10, 6961. https://doi.org/10.3390/app10196961
Vitale AM, Santonocito R, Vergilio G, Marino Gammazza A, Campanella C, Conway de Macario E, Bucchieri F, Macario AJL, Caruso Bavisotto C. Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs. Applied Sciences. 2020; 10(19):6961. https://doi.org/10.3390/app10196961
Chicago/Turabian StyleVitale, Alessandra Maria, Radha Santonocito, Giuseppe Vergilio, Antonella Marino Gammazza, Claudia Campanella, Everly Conway de Macario, Fabio Bucchieri, Alberto J. L. Macario, and Celeste Caruso Bavisotto. 2020. "Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs" Applied Sciences 10, no. 19: 6961. https://doi.org/10.3390/app10196961