Improvement of Soil Microbial Diversity through Sustainable Agricultural Practices and Its Evaluation by -Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety
Abstract
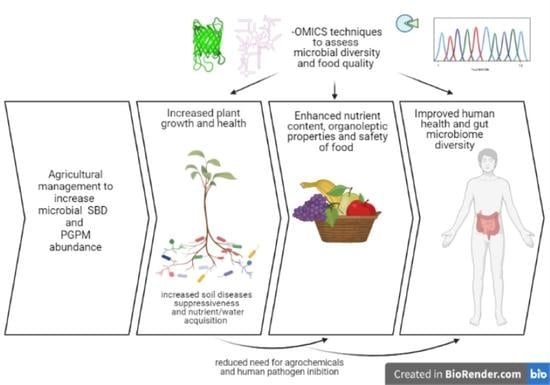
:1. Introduction
2. Using-Omics to Assess Soil Microbial Diversity
2.1. Soil Nucleic Acid High-Throughput Sequencing Technologies
2.2. Soil Metaproteomics
2.3. Soil Enzymomics
2.4. Soil Metabolomics
3. Targeted and Untargeted Approaches to Soil Microbial Diversity Management
| Reported Organisms | Molecular Technique | Inference | Reference |
|---|---|---|---|
| Mycorrhizal fungi (AMF) | Metabolomics | Enhanced plant response to water stress, modulation of oxidative stress conditions and increased production of phytohormone | [74] |
| Endophytic fungi | Metabolomics | Increased plant and fruit yields thanks to improved production of phytohormones | [75] |
| Bacteria | 16S rDNA gene pyrosequencing | Biofertilizers have effects on plant growth, rhizospheric microbes and native microbial communities | [76] |
| Bacteria and fungi | high-throughput pyrosequencing of bacterial 16SV1-V3 and fungal ITS2 of the ribosomal DNA operon | Organic farming increases microbial soil microbial diversity | [77] |
| Rhizospheric Bacteria | high-throughput sequencing of bacterial 16S rDNA gene amplicon | Pepper plants recruit beneficial microbes more efficiently under organic management, thereby increasing soil disease suppression | [78] |
| Bacteria | 16S rDNA gene next generation sequences | Organic farming increases microbial soil microbial diversity | [79] |
| Bacteria | Metaproteomic associated with 16S rDNA genes sequencing | Biostimulant enhances the beneficial activity of microbes on plant growth | [61] |
| Bacteria and archaea | RT-PCR and pyrosequencing of 16S rDNA genes | No differences between conventional and organic farming on the composition of microbial communities | [80] |
| Rhizospheric bacteria | 16S rDNA V3 region gene sequence | Biostimulant increases rare bacterial taxa, some of which involved in plant growth and pathogen resistance | [81] |
| Bacteria | 16S rDNA gene sequences | Tritordeum cv. hire beneficial microbes more efficiently under organic management | [82] |
| Bacteria | Amplification of 16S rDNA V3-V4 regions and high-throughput sequencing | Rotary tillage and straw returning increase bacterial diversity | [83] |
- An untargeted approach based on agricultural practices (increased landscape diversity, complexity and connectivity between the natural ecosystem and agricultural fields; low-input management practices, such as organic farming; strategic crop rotation; intercropping; cover crops; agroforestry; minimum tillage with residue retention; green manures) applied to soil in order to increase biodiversity in favor of multifunctionality, resilience and adaptability to environmental changes [9]; it is a long-term strategy that takes into consideration the complexity, complementarity and self-regulation of soil biota.
- A targeted approach based on the knowledge of soil–plant interactions for specific crops which considers the biotechnological application of microorganisms such as biofertilizers, biostimulants, biopesticides or bioherbicides. This strategy still requires further investigation since the effects of applying specific microorganisms to a native microbial community is not fully understood yet.
3.1. Untargeted Approach
3.1.1. Organic Farming
3.1.2. Conservation Agriculture
3.2. Targeted Approach
4. Implications of Soil Biodiversity for Nutrition and Food Safety
4.1. Food Nutritional Properties
4.2. Food Safety
5. How Beneficial Soil Microbes, Food and Gut Are Interconnected
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bouma, J.; Montanarella, L.; Evanylo, G. The challenge for the soil science community to contribute to the implementation of the UN sustainable development goals. Soil Use Manag. 2019, 35, 538–546. [Google Scholar] [CrossRef]
- Doran, J.W.; Zeiss, M.R. Soil health and sustainability: Managing the biotic component of soil quality. Appl. Soil Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Van Leeuwen, J.P.; Creamer, R.E.; Cluzeau, D.; Debeljak, M.; Gatti, F.; Henriksen, C.B.; Kuzmanovski, V.; Menta, C.; Pérès, G.; Picaud, C.; et al. Modeling of soil functions for assessing soil quality: Soil biodiversity and habitat provisioning. Front. Environ. Sci. 2019, 7, 113. [Google Scholar] [CrossRef]
- Kibblewhite, M.G.; Ritz, K.; Swift, M.J. Soil health in agricultural systems. Philos. Trans. R. Soc. 2008, 363, 685–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrarini, A.; Bini, C.; Amaducci, S. Soil and ecosystem services: Current knowledge and evidences from Italian case studies. Appl. Soil Ecol. 2018, 123, 693–698. [Google Scholar] [CrossRef]
- Karlen, D.L.; Veum, K.S.; Sudduth, K.A.; Obrycki, J.F.; Nunes, M.R. Soil health assessment: Past accomplishments, current activities, and future opportunities. Soil Tillage Res. 2019, 195, 104365. [Google Scholar] [CrossRef]
- Stone, D.; Blomkvist, P.; Hendriksen, N.B.; Bonkowski, M.; Jørgensen, H.B.; Carvalho, F.; Dunbar, M.B.; Gardi, C.; Geisen, S.; Griffiths, R.; et al. A method of establishing a transect for biodiversity and ecosystem function monitoring across Europe. Appl. Soil Ecol. 2016, 97, 3–11. [Google Scholar] [CrossRef]
- EEA. 2016. Available online: Eea.Europa.Eu/Data-and-Maps/Data/Biogeographical-Regions-Europe-3 (accessed on 17 May 2021).
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [Green Version]
- European Environment Agency. Land and Soil in Europe: Why We Need to Use These Vital and Finite Resources Sustainably; Publications Office: Luxemburg, 2019. [Google Scholar]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Nat. Acad. Sci. 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [Green Version]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evolut. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812. [Google Scholar] [CrossRef] [Green Version]
- De Vries, F.T.; Wallenstein, M.D. Below-ground connections underlying above-ground food production: A framework for optimising ecological connections in the rhizosphere. J. Ecol. 2017, 105, 913–920. [Google Scholar] [CrossRef] [Green Version]
- Kaul, S.; Gupta, S.; Sharma, T.; Dhar, M.K. Unfolding the Role of Rhizomicrobiome Toward Sustainable Agriculture. In Root Biology; Spinger: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Hirt, H. Healthy soils for healthy plants for healthy humans: How beneficial microbes in the soil, food and gut are interconnected and how agriculture can contribute to human health. EMBO Rep. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hidalgo, P.; Maymon, M.; Pule-Meulenberg, F.; Hirsch, A.M. Engineering root microbiomes for healthier crops and soils using beneficial, environmentally safe bacteria. Can. J. Microbiol. 2019, 65, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, S.; Wang, J.; Wei, G.; Chen, W.; Lu, Y. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere 2019, 235, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2017, 68, 12–26. [Google Scholar] [CrossRef]
- Bouchez, T.; Blieux, A.L.; Dequiedt, S.; Domaizon, I.; Dufresne, A.; Ferreira, S.; Godon, J.J.; Hellal, J.; Joulian, C.; Quaiser, A.; et al. Molecular microbiology methods for environmental diagnosis. Environ. Chem. Lett. 2016, 14, 423–441. [Google Scholar] [CrossRef]
- Roy, K.; Ghosh, D.; DeBruyn, J.M.; Dasgupta, T.; Wommack, K.E.; Liang, X.; Wagner, R.E.; Radosevich, M. Temporal dynamics of soil virus and bacterial populations in agricultural and early plant successional soils. Front. Microbiol. 2020, 11, 1494. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhuang, J.; Löffler, F.E.; Zhang, Y.; DeBruyn, J.M.; Wilhelm, S.W.; Schaeffer, S.M.; Radosevich, M. Viral and bacterial community responses to stimulated Fe(III)-Bioreduction during simulated subsurface bioremediation. Environ. Microbiol. 2019, 21, 2043–2055. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Vogel, T.M.; Hirsch, P.R.; Simonet, P.; Jansson, J.K.; Tiedje, J.M.; van Elsas, J.D.; Nalin, R.; Philippot, L.; Bailey, M.J. Advantages of the metagenomic approach for soil exploration: Reply from vogel et al. Nat. Rev. Microbiol. 2009, 7, 756–757. [Google Scholar] [CrossRef] [Green Version]
- Baldrian, P. The known and the unknown in soil microbial ecology. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef]
- Bell, T. Next-generation experiments linking community structure and ecosystem functioning. Environ. Microbiol. Rep. 2019, 11, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I. Putting science back into microbial ecology: A question of approach. Philos. Trans. R. Soc. B 2020, 375, 20190240. [Google Scholar] [CrossRef] [Green Version]
- Sala-Comorera, L.; Caudet-Segarra, L.; Galofré, B.; Lucena, F.; Blanch, A.R.; García-Aljaro, C. Unravelling the composition of tap and mineral water microbiota: Divergences between next-generation sequencing techniques and culture-based methods. Int. J. Food Microbiol. 2020, 334, 108850. [Google Scholar] [CrossRef]
- Mishra, M.; Singh, S.K.; Kumar, A. Chapter 32-Role of omics approaches in microbial bioremediation. In Microbe Mediated Remediation of Environmental Contaminants; Kumar, A., Singh, V.K., Singh, P., Mishra, V.K., Eds.; Woodhead Publishing: Sawston, UK, 2021; pp. 435–445. ISBN 978-0-12-821199-1. [Google Scholar]
- Geisen, S.; Briones, M.J.I.; Gan, H.; Behan-Pelletier, V.M.; Friman, V.-P.; de Groot, G.A.; Hannula, S.E.; Lindo, Z.; Philippot, L.; Tiunov, A.V.; et al. A methodological framework to embrace soil biodiversity. Soil Biol. Biochem. 2019, 136, 107536. [Google Scholar] [CrossRef]
- Chandran, H.; Meena, M.; Sharma, K. Microbial biodiversity and bioremediation assessment through omics approaches. Front. Environ. Chem. 2020, 1, 9. [Google Scholar] [CrossRef]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.; Gohl, D.M.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the information content of shallow shotgun metagenomics. mSystems 2018, 3, e00069-18. [Google Scholar] [CrossRef] [Green Version]
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Starke, R.; Jehmlich, N.; Bastida, F. Using proteins to study how microbes contribute to soil ecosystem services: The current state and future perspectives of soil metaproteomics. J. Proteom. 2019, 198, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Abiraami, T.V.; Singh, S.; Nain, L. Soil metaproteomics as a tool for monitoring functional microbial communities: Promises and challenges. Rev. Environ. Sci. BioTechnol. 2020, 19, 73–102. [Google Scholar] [CrossRef]
- Heyer, R.; Schallert, K.; Büdel, A.; Zoun, R.; Dorl, S.; Behne, A.; Kohrs, F.; Püttker, S.; Siewert, C.; Muth, T.; et al. A Robust and universal metaproteomics workflow for research studies and routine diagnostics within 24 h using phenol extraction, FASP digest, and the metaproteomeanalyzer. Front. Microbiol. 2019, 10, 1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A Decade of Metaproteomics: Where we stand and what the future holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef] [Green Version]
- Zampieri, E.; Chiapello, M.; Daghino, S.; Bonfante, P.; Mello, A. Soil metaproteomics reveals an inter-kingdom stress response to the presence of black truffles. Sci. Rep. 2016, 6, 25773. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, T. Potential of enzymomics methodologies to characterize disease-related protein functions. Chem. Pharm. Bull. 2017, 65, 605–610. [Google Scholar] [CrossRef]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E. The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Ch. Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef]
- Kallenbach, C.M.; Frey, S.D.; Grandy, A.S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 2016, 7, 13630. [Google Scholar] [CrossRef]
- Smith, P.; Cotrufo, M.F.; Rumpel, C.; Paustian, K.; Kuikman, P.; Elliott, J.A. Biogeochemical Cycles and biodiversity as key drivers of ecosystem services provided by soils. SOIL Discuss. 2015, 2, 537–586. [Google Scholar]
- Moscatelli, M.C.; Secondi, L.; Marabottini, R.; Papp, R.; Stazi, S.R.; Mania, E.; Marinari, S. Assessment of soil microbial functional diversity: Land use and soil properties affect CLPP-microresp and enzymes responses. Pedobiologia 2018, 66, 36–42. [Google Scholar] [CrossRef]
- Ferrarini, A.; Martani, E.; Fornasier, F.; Amaducci, S. High C input by perennial energy crops boosts belowground functioning and increases soil organic P content. Agric. Ecosyst. Environ. 2021, 308, 107247. [Google Scholar] [CrossRef]
- Marx, M.-C.; Wood, M.; Jarvis, S.C. A Microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol. Biochem. 2001, 33, 1633–1640. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Weintraub, M.N. Emerging tools for measuring and modeling the in situ activity of soil extracellular enzymes. Soil Biol. Biochem. 2008, 40, 2098–2106. [Google Scholar] [CrossRef]
- Trasar-Cepeda, C.; Leirós, M.C.; Gil-Sotres, F. Hydrolytic enzyme activities in agricultural and forest soils. Some implications for their use as indicators of soil quality. Soil Biol. Biochem. 2008, 40, 2146–2155. [Google Scholar] [CrossRef] [Green Version]
- Baldrian, P. Distribution of extracellular enzymes in soils: Spatial heterogeneity and determining factors at various scales. Soil Sci. Soc. Am. J. 2014, 78, 11–18. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Trivedi, C.; Hu, H.; Anderson, I.C.; Jeffries, T.C.; Zhou, J.; Singh, B.K. Microbial regulation of the soil carbon cycle: Evidence from gene–enzyme relationships. ISME J. 2016, 10, 2593–2604. [Google Scholar] [CrossRef] [Green Version]
- Fornasier, F.; Dudal, Y.; Quiquampoix, H. Enzyme Extraction from Soil. In Methods of Soil Enzymology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; pp. 371–383. ISBN 978-0-89118-858-2. [Google Scholar]
- Margon, A.; Fornasier, F. Determining soil enzyme location and related kinetics using rapid fumigation and high-yield extraction. Soil Biol. Biochem. 2008, 40, 2178–2181. [Google Scholar] [CrossRef]
- Fornasier, F.; Ascher, J.; Ceccherini, M.T.; Tomat, E.; Pietramellara, G. A simplified rapid, low-cost and versatile DNA-based assessment of soil microbial biomass. Ecol. Indic. 2014, 45, 75–82. [Google Scholar] [CrossRef]
- Bragato, G.; Fornasier, F.; Brus, D.J. Characterization of soil fertility and soil biodiversity with DsDNA as a covariate in a regression estimator for mean microbial biomass C: Soil DsDNA as a covariate for microbial biomass, C. Eur. J. Soil Sci. 2016, 67, 827–834. [Google Scholar] [CrossRef]
- Bardelli, T.; Ascher-Jenull, J.; Burkia Stocker, E.; Fornasier, F.; Arfaioli, P.; Fravolini, G.; Alves Medeiros, L.R.; Egli, M.; Pietramellara, G.; Insam, H.; et al. Impact of slope exposure on chemical and microbiological properties of norway spruce deadwood and underlying soil during early stages of decomposition in the italian alps. Catena 2018, 167, 100–115. [Google Scholar] [CrossRef]
- Cowie, A.L.; Lonergan, V.E.; Rabbi, S.M.F.; Fornasier, F.; Macdonald, C.; Harden, S.; Kawasaki, A.; Singh, B.K. Impact of carbon farming practices on soil carbon in northern New South Wales. Soil Res. 2013, 51, 707. [Google Scholar] [CrossRef]
- Karas, P.A.; Baguelin, C.; Pertile, G.; Papadopoulou, E.S.; Nikolaki, S.; Storck, V.; Ferrari, F.; Trevisan, M.; Ferrarini, A.; Fornasier, F.; et al. Assessment of the impact of three pesticides on microbial dynamics and functions in a lab-to-field experimental approach. Sci. Total Environ. 2018, 637–638, 636–646. [Google Scholar] [CrossRef]
- Mattarozzi, M.; Di Zinno, J.; Montanini, B.; Manfredi, M.; Marengo, E.; Fornasier, F.; Ferrarini, A.; Careri, M.; Visioli, G. Biostimulants applied to maize seeds modulate the enzymatic activity and metaproteome of the rhizosphere. Appl. Soil Ecol. 2020, 148, 103480. [Google Scholar] [CrossRef]
- Ferrarini, A.; Fracasso, A.; Spini, G.; Fornasier, F.; Taskin, E.; Fontanella, M.C.; Beone, G.M.; Amaducci, S.; Puglisi, E. Bioaugmented Phytoremediation of Metal-Contaminated Soils and Sediments by Hemp and Giant Reed. Front. Microbiol. 2021, 12, 645893. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldridge, B.B.; Rhee, K.Y. Microbial metabolomics: Innovation, application, insight. Curr. Opin. Microbiol. 2014, 19, 90–96. [Google Scholar] [CrossRef]
- Planchamp, C.; Glauser, G.; Mauch-Mani, B. Root inoculation with pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 2015, 5, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pétriacq, P.; Williams, A.; Cotton, A.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. 2017, 92, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Greening, C.; Rattray, J.E.; Chakraborty, A.; Chuvochina, M.; Mayumi, D.; Dolfing, J.; Li, C.; Brooks, J.M.; Bernard, B.B.; et al. Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat. Commun. 2019, 10, 1816. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, H.; White, J.C.; Chen, X.; Li, H.; Qu, X.; Ji, R. Metabolomics reveals that engineered nanomaterial exposure in soil alters both soil rhizosphere metabolite profiles and maize metabolic pathways. Environ. Sci. Nano 2019, 6, 1716–1727. [Google Scholar] [CrossRef]
- Bommarco, R.; Kleijn, D.; Potts, S.G. Ecological intensification: Harnessing ecosystem services for food security. Trends Ecol. Evol. 2013, 28, 230–238. [Google Scholar] [CrossRef]
- Tittonell, P. Ecological intensification of agriculture—Sustainable by nature. Curr. Opin. Environ. Sustain. 2014, 8, 53–61. [Google Scholar] [CrossRef]
- Rey, F.; Cécillon, L.; Cordonnier, T.; Jaunatre, R.; Loucougaray, G. Integrating ecological engineering and ecological intensification from management practices to ecosystem services into a generic framework: A review. Agron. Sustain. Dev. 2015, 35, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Kleijn, D.; Bommarco, R.; Fijen, T.P.M.; Garibaldi, L.A.; Potts, S.G.; van der Putten, W.H. Ecological intensification: Bridging the gap between science and practice. Trends Ecol. Evol. 2018, 34, 154–166. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, L.; Carletti, P.; Badeck, F.W.; Rizza, F.; Morcia, C.; Ghizzoni, R.; Rouphael, Y.; Colla, G.; Terzi, V.; Lucini, L. Metabolomic responses triggered by arbuscular mycorrhiza enhance tolerance to water stress in wheat cultivars. Plant Physiol. Biochem. 2019, 137, 203–212. [Google Scholar] [CrossRef]
- Bonini, P.; Rouphael, Y.; Moreno, M.B.M.; Lee, B.; Cardarelli, M.; Erice, G.; Cirino, V.; Lucini, L.; Colla, G. Microbial-based biostimulant enhances sweet pepper performance by metabolic reprogramming of phytohormone profile and secondary plant metabolism. Front. Plant Sci. 2020, 11, 567388. [Google Scholar] [CrossRef]
- Dal Cortivo, C.; Ferrari, M.; Visioli, G.; Lauro, M.; Fornasier, F.; Barion, G.; Panozzo, A.; Vamerali, T. Effects of seed-applied biofertilizers on rhizosphere biodiversity and growth of common wheat (Triticum Aestivum, L.) in the field. Front. Plant Sci. 2020, 11, 72. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Cai, X.; Gong, J.; Xu, T.; Ding, G.; Li, J. Long-term organic farming manipulated rhizospheric microbiome and bacillus antagonism against pepper blight (Phytophthora Capsici). Front. Microbiol. 2019, 10, 342. [Google Scholar] [CrossRef]
- Lupatini, M.; Korthals, G.W.; de Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Pershina, E.; Valkonen, J.; Kurki, P.; Ivanova, E.; Chirak, E.; Korvigo, I.; Provorov, N.; Andronov, E. Comparative analysis of prokaryotic communities associated with organic and conventional farming systems. PLoS ONE 2015, 10, e0145072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visioli, G.; Sanangelantoni, A.M.; Vamerali, T.; Dal Cortivo, C.; Blandino, M. 16S RDNA profiling to reveal the influence of seed-applied biostimulants on the rhizosphere of young maize plants. Molecules 2018, 23, 1461. [Google Scholar] [CrossRef] [Green Version]
- Visioli, G.; Lauro, M.; Vamerali, T.; Dal Cortivo, C.; Panozzo, A.; Folloni, S.; Piazza, C.; Ranieri, R. A Comparative study of organic and conventional management on the rhizosphere microbiome, growth and grain quality traits of tritordeum. Agronomy 2020, 10, 1717. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, P.; He, L.; Gao, X.; Li, W.; Zhou, Y.; Li, Z.; Li, H.; Yang, L. Effects of tillage managements and maize straw returning on soil microbiome using 16S RDNA sequencing. J. Integr. Plant Biol. 2019, 61, 765–777. [Google Scholar] [CrossRef] [Green Version]
- EC (European Commission). 2016. Organic Certification [on Line], European Commission, DG Agriculture and Rural Development, Unit Agricultural Modelling and Outlook, Brussels. Available online: Ec.Europa.Eu/Agriculture/Organic/Organicfarming/What-Is-Organic-Farming/Organiccertification_en (accessed on 24 June 2020).
- USDA (United States Department of Agriculture). 2016. Organic Regulations. Available online: Ams.Usda.Gov/Rulesregulations/Organic (accessed on 24 June 2020).
- Dal Ferro, N.; Zanin, G.; Borin, M. Crop yield and energy use in organic and conventional farming: A case study in North-East Italy. Eur. J. Agron. 2017, 86, 37–47. [Google Scholar] [CrossRef]
- Gomiero, T. Food quality assessment in organic vs. conventional agricultural produce: Findings and issues. Appl. Soil Ecol. 2018, 123, 714–728. [Google Scholar] [CrossRef]
- Schrama, M.; de Haan, J.J.; Kroonen, M.; Verstegen, H.; Van der Putten, W.H. Crop yield gap and stability in organic and conventional farming systems. Agric. Ecosyst. Environ. 2018, 256, 123–130. [Google Scholar] [CrossRef]
- Li, M.; Peterson, C.A.; Tautges, N.E.; Scow, K.M.; Gaudin, A.C.M. Yields and resilience outcomes of organic, cover crop, and conventional practices in a mediterranean climate. Sci. Rep. 2019, 9, 12283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson, J.; Ahnstrom, J.; Weibull, A.-C. The effects of organic agriculture on biodiversity and abundance: A meta-analysis. J. Appl. Ecol. 2005, 42, 261–269. [Google Scholar] [CrossRef]
- Lori, M.; Symnaczik, S.; Mader, P.; De Deyn, G.; Gattinger, A. Organic farming enhances soil microbial abundance and activity—A meta-analysis and meta-regression. PLoS ONE 2017, 12, 1–25. [Google Scholar]
- Xia, Y.; Sahib, M.R.; Amna, A.; Opiyo, S.O.; Zhao, Z.; Gao, Y.G. Culturable endophytic fungal communities associated with plants in organic and conventional farming systems and their effects on plant growth. Sci. Rep. 2019, 9, 1669. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.S.; Panwar, J.; Meena, H.N.; Thirumalaisamy, P.P.; Meena, R.L. Developing Disease-Suppressive Soil Through Agronomic Management. In Organic Amendments and Soil Suppressiveness in Plant Disease Management; Meghvansi, M.K., Varma, A., Eds.; Soil Biology; Springer International Publishing: Cham, Switzerland, 2015; Volume 46, pp. 61–94. ISBN 978-3-319-23074-0. [Google Scholar]
- Nielsen, U.N.; Wall, D.H.; Six, J. Soil biodiversity and the environment. Annu. Rev. Environ. Resour. 2015, 40, 63–90. [Google Scholar] [CrossRef]
- Meena, V.S.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A. Agriculturally Important Microbes for Sustainable Agriculture: Volume 2: Applications in Crop Production and Protection; Springer Nature Singapore Pte Ltd.: Singapore, 2017; p. 189721. ISBN 978-981-10-5342-9. [Google Scholar]
- Lal, R.; Stewart, B.A. Soil Degradation and Restoration in Africa; CRC Press: Boca Raton, FL, USA, 2019; ISBN 978-1-351-59330-4. [Google Scholar]
- Barrios, E.; Sileshi, G.W.; Shepherd, K.; Sinclair, F. Agroforestry and Soil Health: Linking Trees, Soil Biota, and Ecosystem Services. In Soil Ecology and Ecosystem Services; Oxford University Press: Oxford, UK, 2012; pp. 315–329. [Google Scholar]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.-J. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Kong, A.Y.Y.; Six, J. Microbial community assimilation of cover crop rhizodeposition within soil microenvironments in alternative and conventional cropping systems. Plant Soil 2012, 356, 315–330. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Rivera-Orduña, F.N.; Patiño-Zúñiga, L.; Vila-Sanjurjo, A.; Crossa, J.; Govaerts, B.; Dendooven, L. Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl. Environ. Microbiol. 2010, 76, 3685–3691. [Google Scholar] [CrossRef] [Green Version]
- Guerrieri, M.C.; Fanfoni, E.; Fiorini, A.; Trevisan, M.; Puglisi, E. Isolation and screening of extracellular PGPR from the rhizosphere of tomato plants after long-term reduced tillage and cover crops. Plants 2020, 9, 668. [Google Scholar] [CrossRef]
- Palm, C.; Blanco-Canqui, H.; DeClerck, F.; Gatere, L.; Grace, P. Conservation agriculture and ecosystem services: An overview. Agric. Ecosyst. Environ. 2014, 187, 87–105. [Google Scholar] [CrossRef] [Green Version]
- Pittelkow, C.M.; Liang, X.; Linquist, B.A.; van Groenigen, K.J.; Lee, J.; Lundy, M.E.; van Gestel, N.; Six, J.; Venterea, R.T.; van Kessel, C. Productivity limits and potentials of the principles of conservation agriculture. Nature 2015, 517, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Powlson, D.S.; Stirling, C.M.; Jat, M.L.; Gerard, B.G.; Palm, C.A.; Sanchez, P.A.; Cassman, K.G. Limited potential of no-till agriculture for climate change mitigation. Nat. Clim. Ch. 2014, 4, 678–683. [Google Scholar] [CrossRef]
- Yadav, A.N.; Kumar, R.; Kumar, S.; Kumar, V. Beneficial microbiomes: Biodiversity and potential biotechnological applications for sustainable agriculture and human health. J. App. Biol. Biotech. 2017, 5, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Esitken, A.; Yildiz, H.E.; Ercisli, S.; Figen Donmez, M.; Turan, M.; Gunes, A. Effects of plant growth promoting bacteria (PGPB) on yield, growth and nutrient contents of organically grown strawberry. Sci. Hortic. 2010, 124, 62–66. [Google Scholar] [CrossRef]
- Gianinazzi, S.; Gollotte, A.; Binet, M.-N.; van Tuinen, D.; Redecker, D.; Wipf, D. Agroecology: The key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 2010, 20, 519–530. [Google Scholar] [CrossRef]
- O’Callaghan, M. Microbial inoculation of seed for improved crop performance: Issues and opportunities. Appl. Microbiol. Biotechnol. 2016, 100, 5729–5746. [Google Scholar] [CrossRef] [PubMed]
- Preininger, C.; Sauer, U.; Bejarano, A.; Berninger, T. Concepts and applications of foliar spray for microbial inoculants. Appl. Microbiol. Biotechnol. 2018, 102, 7265–7282. [Google Scholar] [CrossRef]
- Bertola, M.; Mattarozzi, M.; Sanangelantoni, A.M.; Careri, M.; Visioli, G. PGPB colonizing three-year biochar-amended soil: Towards biochar-mediated biofertilization. J. Soil Sci. Plant Nutr. 2019, 19, 841–850. [Google Scholar] [CrossRef]
- Egamberdiyeva, D. The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl. Soil Ecol. 2007, 36, 184–189. [Google Scholar] [CrossRef]
- Fiorentino, N.; Ventorino, V.; Woo, S.L.; Pepe, O.; De Rosa, A.; Gioia, L.; Romano, I.; Lombardi, N.; Napolitano, M.; Colla, G.; et al. Trichoderma-based biostimulants modulate rhizosphere microbial populations and improve N uptake efficiency, yield, and nutritional quality of leafy vegetables. Front. Plant Sci. 2018, 9, 743. [Google Scholar] [CrossRef] [Green Version]
- Dal Cortivo, C.; Barion, G.; Visioli, G.; Mattarozzi, M.; Mosca, G.; Vamerali, T. Increased root growth and nitrogen accumulation in common wheat following PGPR inoculation: Assessment of plant-microbe interactions by ESEM. Agric. Ecosyst. Environ. 2017, 247, 396–408. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Plant-Microbe Interactions in Adaptation of Agricultural Crops to Abiotic Stress Conditions. In Probiotics and Plant Health; Springer: Singapore, 2017. [Google Scholar] [CrossRef]
- Paul, K.; Sorrentino, M.; Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Miras Moreno, M.B.; Reynaud, H.; Canaguier, R.; Trtílek, M.; et al. A combined phenotypic and metabolomic approach for elucidating the biostimulant action of a plant-derived protein hydrolysate on tomato grown under limited water availability. Front. Plant Sci. 2019, 10, 493. [Google Scholar] [CrossRef]
- Camprubi, A.; Solari, J.; Bonini, P.; Garcia-Figueres, F.; Colosimo, F.; Cirino, V.; Lucini, L.; Calvet, C. Plant performance and metabolomic profile of loquat in response to mycorrhizal inoculation, armillaria mellea and their interaction. Agronomy 2020, 10, 899. [Google Scholar] [CrossRef]
- Kokkoris, V.; Vukicevich, E.; Richards, A.; Thomsen, C.; Hart, M.M. Challenges using droplet digital PCR for environmental samples. Appl. Microbiol. 2021, 1, 7. [Google Scholar] [CrossRef]
- Rilling, J.I.; Acuña, J.J.; Nannipieri, P.; Cassan, F.; Maruyama, F.; Jorquera, M.A. Current opinion and perspectives on the methods for tracking and monitoring plant growth‒promoting bacteria. Soil Biol. Biochem. 2019, 130, 205–219. [Google Scholar] [CrossRef]
- Romano, I.; Ventorino, V.; Pepe, O. Effectiveness of Plant Beneficial Microbes: Overview of the Methodological Approaches for the Assessment of Root Colonization and Persistence. Front. Plant Sci. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabacchioni, S.; Passato, S.; Ambrosino, P.; Huang, L.; Caldara, M.; Cantale, C.; Hett, J.; Del Fiore, A.; Fiore, A.; Schlüter, A.; et al. Identification of Beneficial Microbial Consortia and Bioactive Compounds with Potential as Plant Biostimulants for a Sustainable Agriculture. Microorganisms 2021, 9, 426. [Google Scholar] [CrossRef]
- Lal, R. Soil degradation as a reason for inadequate human nutrition. Food Sec. 2009, 1, 45–57. [Google Scholar] [CrossRef]
- Brevik, E.C.; Sauer, T.J. The past, present, and future of soils and human health studies. Soil 2015, 1, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Rojas, R.V.; Achouri, M.; Maroulis, J.; Caon, L. Healthy soils: A prerequisite for sustainable food security. Environ. Earth Sci. 2016, 75, 180. [Google Scholar] [CrossRef]
- Kaur, T.; Rana, K.L.; Kour, D.; Sheikh, I.; Yadav, N.; Kumar, V.; Yadav, A.N.; Dhaliwal, H.S.; Saxena, A.K. Microbe-mediated biofortification for micronutrients: Present status and future challenges. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–17. ISBN 978-0-12-820528-0. [Google Scholar]
- El Mujtar, V.; Muñoz, N.; Prack Mc Cormick, B.; Pulleman, M.; Tittonell, P. Role and management of soil biodiversity for food security and nutrition; where do we stand? Glob. Food Secur. 2019, 20, 132–144. [Google Scholar] [CrossRef]
- Baslam, M.; Garmendia, I.; Goicoechea, N. Arbuscular mycorrhizal fungi (AMF) improved growth and nutritional quality of greenhouse-grown lettuce. J. Agric. Food Chem. 2011, 59, 5504–5515. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Cantamessa, S.; Massa, N.; Manassero, P.; Marsano, F.; Copetta, A.; Lingua, G.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads improve yield, quality and nutritional value of tomato: A field study. Mycorrhiza 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Giovannetti, M.; Avio, L.; Barale, R.; Ceccarelli, N.; Cristofani, R.; Iezzi, A.; Mignolli, F.; Picciarelli, P.; Pinto, B.; Reali, D.; et al. Nutraceutical value and safety of tomato fruits produced by mycorrhizal plants. Br. J. Nutr. 2012, 107, 242–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalid, M.; Hassani, D.; Bilal, M.; Asad, F.; Huang, D. Influence of bio-fertilizer containing beneficial fungi and rhizospheric bacteria on health promoting compounds and antioxidant activity of spinacia Oleracea, L. Bot. Stud. 2017, 58, 35. [Google Scholar] [CrossRef] [Green Version]
- De Silva, A.; Patterson, K.; Rothrock, C.; Moore, J. Growth promotion of highbush blueberry by fungal and bacterial inoculants. HortScience 2000, 35, 1228–1230. [Google Scholar] [CrossRef] [Green Version]
- Lingua, G.; Bona, E.; Manassero, P.; Marsano, F.; Todeschini, V.; Cantamessa, S.; Copetta, A.; D’Agostino, G.; Gamalero, E.; Berta, G. Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads increases anthocyanin concentration in strawberry fruits (Fragaria × Ananassa Var. Selva) in conditions of reduced fertilization. IJMS 2013, 14, 16207–16225. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Joshi, M.; Prasanna, R.; Shivay, Y.S.; Nain, L. Biofortification of wheat through inoculation of plant growth promoting rhizobacteria and cyanobacteria. Eur. J. Soil Biol. 2012, 50, 118–126. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Zhang, X.; Dong, L.; Zhang, J.; Wei, Y.; Feng, Y.; Lu, L. Improved plant growth and Zn accumulation in grains of rice (Oryza Sativa, L.) by inoculation of endophytic microbes isolated from a Zn Hyperaccumulator, Sedum Alfredii, H.J. Agric. Food Chem. 2014, 62, 1783–1791. [Google Scholar] [CrossRef]
- Yaseen, M. Microbial assisted foliar feeding of micronutrients enhance growth, yield and biofortification of wheat. IJAB 2018, 353–360. [Google Scholar] [CrossRef]
- Reeve, J.R.; Hoagland, L.A.; Villalba, J.J.; Carr, P.M.; Atucha, A.; Cambardella, C.; Davis, D.R.; Delate, K. Organic Farming, Soil Health, and Food Quality: Considering Possible Links. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2016; Volume 137, pp. 319–367. ISBN 978-0-12-804692-0. [Google Scholar]
- Avio, L.; Turrini, A.; Giovannetti, M.; Sbrana, C. Designing the ideotype mycorrhizal symbionts for the production of healthy food. Front. Plant Sci. 2018, 9, 1089. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Babalola, O.O. Microbial inoculants for improving crop quality and human health in Africa. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef] [Green Version]
- Kolega, S.; Miras-Moreno, B.; Buffagni, V.; Lucini, L.; Valentinuzzi, F.; Maver, M.; Mimmo, T.; Trevisan, M.; Pii, Y.; Cesco, S. Nutraceutical profiles of two hydroponically grown sweet basil cultivars as affected by the composition of the nutrient solution and the inoculation with azospirillum brasilense. Front. Plant Sci. 2020, 11, 596000. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Fausto, C.; Mininni, A.N.; Dichio, B.; Lucini, L. Soil management type differentially modulates the metabolomic profile of olive xylem sap. Plant Physiol. Biochem. 2019, 139, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Armesto, J.; Rocchetti, G.; Senizza, B.; Pateiro, M.; Barba, F.J.; Domínguez, R.; Lucini, L.; Lorenzo, J.M. Nutritional characterization of butternut squash (Cucurbita Moschata, D.): Effect of variety (Ariel vs. Pluto) and farming type (Conventional vs. Organic). Food Res. Int. 2020, 132, 109052. [Google Scholar] [CrossRef]
- Rocchetti, G.; Braceschi, G.P.; Odello, L.; Bertuzzi, T.; Trevisan, M.; Lucini, L. Identification of markers of sensory quality in ground coffee: An untargeted metabolomics approach. Metabolomics 2020, 16, 127. [Google Scholar] [CrossRef]
- Dal Cortivo, C.; Barion, G.; Ferrari, M.; Visioli, G.; Dramis, L.; Panozzo, A.; Vamerali, T. Effects of field inoculation with VAM and bacteria consortia on root growth and nutrients uptake in common wheat. Sustainability 2018, 10, 3286. [Google Scholar] [CrossRef] [Green Version]
- Rillig, M.C.; Lehmann, A.; Lehmann, J.; Camenzind, T.; Rauh, C. Soil biodiversity effects from field to fork. Trends Plant Sci. 2018, 23, 17–24. [Google Scholar] [CrossRef]
- Torri, L.; Migliorini, P.; Masoero, G. Sensory test vs. electronic nose and/or image analysis of whole bread produced with old and modern wheat varieties adjuvanted by means of the mycorrhizal factor. Food Res. Int. 2013, 54, 1400–1408. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, Q.; Yuan, Y.; Sun, W. Human health risk assessment of heavy metals in soil and food crops in the pearl river delta urban agglomeration of China. Food Chem. 2020, 316, 126213. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.A.; Gregory, P.J. Soil, food security and human health: A review: Soil, food security and human health. Eur. J. Soil Sci. 2015, 66, 257–276. [Google Scholar] [CrossRef]
- Zhuang, P.; McBride, M.B.; Xia, H.; Li, N.; Li, Z. Health risk from heavy metals via consumption of food crops in the vicinity of Dabaoshan Mine, South China. Sci. Total Environ. 2009, 407, 1551–1561. [Google Scholar] [CrossRef]
- Li, H.; Chen, X.W.; Wong, M.H. Arbuscular mycorrhizal fungi reduced the ratios of inorganic/organic arsenic in rice grains. Chemosphere 2016, 145, 224–230. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, Y.; Song, A.; Yu, B.; Zeng, X.; Chen, M.-S.; Yin, H.; Zhang, X.; Sun, B.; Fan, F. Loss of soil microbial diversity may increase insecticide uptake by crop. Agric. Ecosyst. Environ. 2017, 240, 84–91. [Google Scholar] [CrossRef]
- Ismail, Y.; McCormick, S.; Hijri, M. The arbuscular mycorrhizal fungus, glomus irregulare, controls the mycotoxin production of fusarium sambucinum in the pathogenesis of potato. FEMS Microbiol. Lett. 2013, 348, 46–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.S.; Fu, Z.; Reganold, J.P.; Karp, D.S.; Besser, T.E.; Tylianakis, J.M.; Snyder, W.E. Organic Farming Promotes Biotic Resistance to Foodborne Human Pathogens. J. Appl. Ecol. 2019, 56, 1117–1127. [Google Scholar] [CrossRef]
- Semenov, A.V.; van Overbeek, L.; van Bruggen, A.H.C. Percolation and survival of Escherichia Coli O157:H7 and Salmonella Enterica serovar typhimurium in soil amended with contaminated dairy manure or slurry. Appl. Environ. Microbiol. 2009, 75, 3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, G.; Cevallos-Cevallos, J.M.; Vallad, G.E.; van Bruggen, A.H.C. Organically managed soils reduce internal colonization of tomato plants by Salmonella Enterica Serovar Typhimurium. Phytopathology 2013, 103, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Van Bruggen, A.H.C.; Goss, E.M.; Havelaar, A.; van Diepeningen, A.D.; Finckh, M.R.; Morris, J.G. One health-cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health. Sci. Total Environ. 2019, 664, 927–937. [Google Scholar] [CrossRef]
- Ramírez-Puebla, S.T.; Servín-Garcidueñas, L.E.; Jiménez-Marín, B.; Bolaños, L.M.; Rosenblueth, M.; Martínez, J.; Rogel, M.A.; Ormeño-Orrillo, E.; Martínez-Romero, E. Gut and root microbiota commonalities. Appl. Environ. Microbiol. 2013, 79, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Blum, W.E.H.; Zechmeister-Boltenstern, S.; Keiblinger, K.M. Does soil contribute to the human gut microbiome? Microorganisms 2019, 7, 287. [Google Scholar] [CrossRef] [Green Version]
- Grieneisen, L.E.; Charpentier, M.J.E.; Alberts, S.C.; Blekhman, R.; Bradburd, G.; Tung, J.; Archie, E.A. Genes, geology and germs: Gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc. R. Soc. B 2019, 286, 20190431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasnim, N.; Abulizi, N.; Pither, J.; Hart, M.M.; Gibson, D.L. Linking the gut microbial ecosystem with the environment: Does gut health depend on where we live? Front. Microbiol. 2017, 8, 1935. [Google Scholar] [CrossRef]
- Martínez, I.; Stegen, J.C.; Maldonado-Gómez, M.X.; Eren, A.M.; Siba, P.M.; Greenhill, A.R.; Walter, J. The gut microbiota of rural papua new guineans: Composition, diversity patterns, and ecological processes. Cell Rep. 2015, 11, 527–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rook, G.A.W. Review series on helminths, immune modulation and the hygiene hypothesis: The broader implications of the hygiene hypothesis. Immunology 2009, 126, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Ruokolainen, L.; Suomalainen, A.; Sinkko, H.; Karisola, P.; Lehtimäki, J.; Lehto, M.; Hanski, I.; Alenius, H.; Fyhrquist, N. Soil exposure modifies the gut microbiota and supports immune tolerance in a mouse model. J. Allergy Clin. Immunol. 2019, 143, 1198–1206.e12. [Google Scholar] [CrossRef] [Green Version]
- Lowry, C.A.; Hollis, J.H.; de Vries, A.; Pan, B.; Brunet, L.R.; Hunt, J.R.F.; Paton, J.F.R.; van Kampen, E.; Knight, D.M.; Evans, A.K.; et al. Identification of an immune-responsive mesolimbocortical serotonergic system: Potential role in regulation of emotional behavior. Neuroscience 2007, 146, 756–772. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.G.; Martinelli, R.; Besra, G.S.; Illarionov, P.A.; Szatmari, I.; Brazda, P.; Allen, M.A.; Xu, W.; Wang, X.; Nagy, L.; et al. Identification and characterization of a novel anti-inflammatory lipid isolated from mycobacterium vaccae, a soil-derived bacterium with immunoregulatory and stress resilience properties. Psychopharmacology 2019, 236, 1653–1670. [Google Scholar] [CrossRef] [PubMed]
- Van Bruggen, A.H.C.; He, M.M.; Shin, K.; Mai, V.; Jeong, K.C.; Finckh, M.R.; Morris, J.G. Environmental and health effects of the herbicide glyphosate. Sci. Total Environ. 2018, 616–617, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Erisman, J.W. Nature-based agriculture for an adequate human microbiome. Org. Agric. 2020. [Google Scholar] [CrossRef]

| Crop | Rhizosphere Microbes | Plant Tissue | Impact on Food Quality and Nutritional Status | Reference |
|---|---|---|---|---|
| Lettuce | Azotobacter chroococcum and Glomus fasciculatum; G. fasciculatum and Glomus mosseae | Leaves | Increased concentration of total phenolic compounds, anthocyanins, and carotenoids; increased flavonoid content | [126] |
| Tomato | Pseudomonas strains and a mixed mycorrhizal inoculum, under conditions of reduced fertilization | Fruit | Larger and better-quality fruits (concentration of sugars, organic acids, such as citric and malic acid, and vitamin C) | [127] |
| Strawberry | Bacillus, Pseudomonas | Fruit | Increased fruit yield, plant growth, nutrient content (P, Fe, Zn, and vitamin C) | [106] |
| Tomato | AMF G. intraradices | Fruit | Increased plant growth, mineral nutrient content (P, K, Ca, Zn), and enhanced nutritional and nutraceutical value (carotenoids such as lycopene) | [128] |
| Spinach | AMF and bacterial species | Leaves | Higher concentration of total phenolic compounds, flavonoids, and phenolic acid | [129] |
| Highbush blueberry | PGPR (Pseudomonas sp. and Bacillus sp.) and AMF (Gliocladium virens and Trichoderma harzianum) | Leaves | Increased plant growth and enhanced P, Zn and Cu uptake | [130] |
| Strawberry | AMF and selected Pseudomonas strains, under conditions of reduced fertilization | Fruit | Increased concentration of antioxidant molecules (anthocyanins). Yield not affected | [131] |
| Wheat grains | PGPB (Providencia sp.) and cyanobacterial strains (Anabaena sp., Calothrix sp. and Anabaena sp.) | Grain | Increased yield, and micronutrient (Fe, Zn, Cu, Mn) and protein enrichment | [132] |
| Rice grains | Endophytic strains, Burkholderia sp., Sphingomonas sp., Variovorax sp., and Enterobacter sp., | Grain | Improved plant growth, yield and root morphology, increased Zn concentration in shoot and roots | [133] |
| Wheat grains | Endophytic bacteria, Enterobacter sp. and Burkholderia phytofirmans | Grain | Enhanced Fe concentration, plant height, leaf area, spike length, and plant biomass | [134] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertola, M.; Ferrarini, A.; Visioli, G. Improvement of Soil Microbial Diversity through Sustainable Agricultural Practices and Its Evaluation by -Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety. Microorganisms 2021, 9, 1400. https://doi.org/10.3390/microorganisms9071400
Bertola M, Ferrarini A, Visioli G. Improvement of Soil Microbial Diversity through Sustainable Agricultural Practices and Its Evaluation by -Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety. Microorganisms. 2021; 9(7):1400. https://doi.org/10.3390/microorganisms9071400
Chicago/Turabian StyleBertola, Marta, Andrea Ferrarini, and Giovanna Visioli. 2021. "Improvement of Soil Microbial Diversity through Sustainable Agricultural Practices and Its Evaluation by -Omics Approaches: A Perspective for the Environment, Food Quality and Human Safety" Microorganisms 9, no. 7: 1400. https://doi.org/10.3390/microorganisms9071400