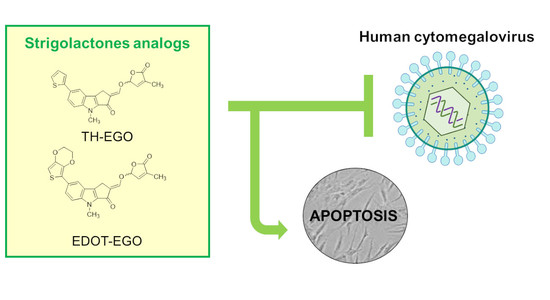
Strigolactone Analogs Are Promising Antiviral Agents for the Treatment of Human Cytomegalovirus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Cells and Viruses
2.3. Antiviral Assays
2.3.1. Virus Yield Reduction Assay
2.3.2. Attachment Assay
2.3.3. Entry Assay
2.4. Cytotoxicity Assay
2.5. Western Blot Analysis
2.6. Apoptosis Detection
2.6.1. Annexin V Analysis
2.6.2. In-Vitro Analysis of Caspase-3 Activity
2.7. Molecular Modeling
2.8. Statistical Analysis
3. Results
3.1. Effects of SL Analogs on HCMV Productive Infection
3.2. The Butenolide Ring Is Critical for the Antiviral Activity of TH-EGO and EDOT-EGO
3.3. TH-EGO and EDOT-EGO Inhibit an Early-Late Event in the HCMV Replication Cycle
3.4. Virolysis as the Mechanism for TH-EGO and EDOT-EGO Antiviral Activity
3.5. Identification of Putative Drug-Binding Target Proteins Using In-Silico Docking Simulations
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, W.D.; Boppana, S.B.; Fowler, K.B.; Kimberlin, D.W.; Lazzarotto, T.; Alain, S.; Daly, K.; Doutré, S.; Gibson, L.; Giles, M.L.; et al. Congenital cytomegalovirus infection in pregnancy and the neonate: Consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect. Dis. 2017, 17, e177–e188. [Google Scholar] [CrossRef]
- Britt, W.J. Maternal Immunity and the Natural History of Congenital Human Cytomegalovirus Infection. Viruses 2018, 10, 405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britt, W.J.; Prichard, M.N. New therapies for human cytomegalovirus infections. Antiviral Res. 2018, 159, 153–174. [Google Scholar] [CrossRef]
- Zwanenburg, B.; Blanco-Ania, D. Strigolactones: New plant hormones in the spotlight. J. Exp. Bot. 2018, 69, 2205–2218. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H.; Prandi, C. Strigolactones: Past, present and future. Planta 2016, 243, 1309. [Google Scholar] [CrossRef] [Green Version]
- Makhzoum, A.; Yousefzadi, M.; Malik, S.; Gantet, P.; Tremouillaux-Guiller, J. Strigolactone biology: Genes, functional genomics, epigenetics and applications. Crit. Rev. Biotechnol. 2017, 37, 151–162. [Google Scholar] [CrossRef]
- Mayzlish-Gati, E.; Laufer, D.; Grivas, C.F.; Shaknof, J.; Sananes, A.; Bier, A.; Ben-Harosh, S.; Belausov, E.; Johnson, M.D.; Artuso, E.; et al. Strigolactone analogs act as new anti-cancer agents in inhibition of breast cancer in xenograft model. Cancer Biol. Ther. 2015, 16, 1682–1688. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.N.; Razvi, S.S.I.; Kuerban, A.; Balamash, K.S.; Al-Bishri, W.M.; Abulnaja, K.O.; Choudhry, H.; Khan, J.A.; Moselhy, S.S.; M, Z.; et al. Strigolactones-a novel class of phytohormones as anti-cancer agents. J. Pestic. Sci. 2018, 43, 168–172. [Google Scholar] [CrossRef]
- Hasan, M.N.; Choudhry, H.; Razvi, S.S.; Moselhy, S.S.; Kumosani, T.A.; Zamzami, M.A.; Omran, Z.; Halwani, M.A.; Al-Babili, S.; Abualnaja, K.O.; et al. Synthetic strigolactone analogues reveal anti-cancer activities on hepatocellular carcinoma cells. Bioorg. Med. Chem. Lett. 2018, 28, 1077–1083. [Google Scholar] [CrossRef] [Green Version]
- Pollock, C.B.; Koltai, H.; Kapulnik, Y.; Prandi, C.; Yarden, R.I. Strigolactones: A novel class of phytohormones that inhibit the growth and survival of breast cancer cells and breast cancer stem-like enriched mammosphere cells. Breast Cancer Res. Treat. 2012, 134, 1041–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollock, C.B.; McDonough, S.; Wang, V.S.; Lee, H.; Ringer, L.; Li, X.; Prandi, C.; Lee, R.J.; Feldman, A.S.; Koltai, H.; et al. Strigolactone analogues induce apoptosis through activation of p38 and the stress response pathway in cancer cell lines and in conditionally reprogrammed primary prostate cancer cells. Oncotarget 2014, 5, 1683–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croglio, M.P.; Haake, J.M.; Ryan, C.P.; Wang, V.S.; Lapier, J.; Schlarbaum, J.P.; Dayani, Y.; Artuso, E.; Prandi, C.; Koltai, H.; et al. Analogs of the novel phytohormone, strigolactone, trigger apoptosis and synergize with PARP inhibitors by inducing DNA damage and inhibiting DNA repair. Oncotarget 2016, 7, 13984–14001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.-X.; Han, Y.-S.; Wang, J.-C.; Yang, H.; Kong, H.; Liu, K.-J.; Chen, S.-Y.; Chen, Y.-R.; Chang, Y.-Q.; Chen, W.-M.; et al. Strigolactones: A plant phytohormone as novel anti-inflammatory agents. Medchemcomm 2018, 9, 181–188. [Google Scholar] [CrossRef]
- Tumer, T.B.; Yılmaz, B.; Ozleyen, A.; Kurt, B.; Tok, T.T.; Taskin, K.M.; Kulabas, S.S. GR24, a synthetic analog of Strigolactones, alleviates inflammation and promotes Nrf2 cytoprotective response: In vitro and in silico evidences. Comput. Biol. Chem. 2018, 76, 179–190. [Google Scholar] [CrossRef]
- Zhu, H.; Shen, Y.; Shenk, T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 1995, 69, 7960–7970. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Alwine, J.C. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 2002, 76, 3731–3738. [Google Scholar] [CrossRef] [Green Version]
- Brune, W.; Andoniou, C.E. Die Another Day: Inhibition of Cell Death Pathways by Cytomegalovirus. Viruses 2017, 9, 249. [Google Scholar] [CrossRef] [Green Version]
- Prandi, C.; Occhiato, E.G.; Tabasso, S.; Bonfante, P.; Novero, M.; Scarpi, D.; Bova, M.E.; Miletto, I. New Potent Fluorescent Analogues of Strigolactones: Synthesis and Biological Activity in Parasitic Weed Germination and Fungal Branching. Eur. J. Org. Chem. 2011, 2011, 3781–3793. [Google Scholar] [CrossRef]
- Dolan, A.; Cunningham, C.; Hector, R.D.; Hassan-Walker, A.F.; Lee, L.; Addison, C.; Dargan, D.J.; McGeoch, D.J.; Gatherer, D.; Emery, V.C.; et al. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004, 85, 1301–1312. [Google Scholar] [CrossRef]
- Grazia Revello, M.; Baldanti, F.; Percivalle, E.; Sarasini, A.; De-Giuli, L.; Genini, E.; Lilleri, D.; Labò, N.; Gerna, G. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J. Gen. Virol. 2001, 82, 1429–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gariano, G.R.; Dell’Oste, V.; Bronzini, M.; Gatti, D.; Luganini, A.; De Andrea, M.; Gribaudo, G.; Gariglio, M.; Landolfo, S. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012, 8, e1002498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luganini, A.; Nicoletto, S.F.; Pizzuto, L.; Pirri, G.; Giuliani, A.; Landolfo, S.; Gribaudo, G. Inhibition of herpes simplex virus type 1 and type 2 infections by peptide-derivatized dendrimers. Antimicrob. Agents Chemother. 2011, 55, 3231–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; von Einem, J.; Marschall, M.; Plachter, B.; Gariglio, M.; De Andrea, M.; Landolfo, S. Regulatory Interaction between the Cellular Restriction Factor IFI16 and Viral pp65 (pUL83) Modulates Viral Gene Expression and IFI16 Protein Stability. J. Virol. 2016, 90, 8238–8250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baroni, M.; Cruciani, G.; Sciabola, S.; Perruccio, F.; Mason, J.S. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): Theory and application. J. Chem. Inf. Model. 2007, 47, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Artuso, E.; Ghibaudi, E.; Lace, B.; Marabello, D.; Vinciguerra, D.; Lombardi, C.; Koltai, H.; Kapulnik, Y.; Novero, M.; Occhiato, E.G.; et al. Stereochemical Assignment of Strigolactone Analogues Confirms Their Selective Biological Activity. J. Nat. Prod. 2015, 78, 2624–2633. [Google Scholar] [CrossRef] [Green Version]
- Kalser, J.; Adler, B.; Mach, M.; Kropff, B.; Puchhammer-Stöckl, E.; Görzer, I. Differences in Growth Properties among Two Human Cytomegalovirus Glycoprotein O Genotypes. Front. Microbiol. 2017, 8, 1609. [Google Scholar] [CrossRef]
- Collins-McMillen, D.; Chesnokova, L.; Lee, B.-J.; Fulkerson, H.L.; Brooks, R.; Mosher, B.S.; Yurochko, A.D. HCMV Infection and Apoptosis: How Do Monocytes Survive HCMV Infection? Viruses 2018, 10, 533. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.R.; Sun, L. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 1997, 71, 2970–2979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koltai, H.; Prandi, C. Strigolactones—Biology and Applications; Springer International Publishing: New York City, NY, USA, 2019; ISBN 978-3-030-12152-5. [Google Scholar]
- Spyrakis, F.; Felici, P.; Bayden, A.S.; Salsi, E.; Miggiano, R.; Kellogg, G.E.; Cozzini, P.; Cook, P.F.; Mozzarelli, A.; Campanini, B. Fine tuning of the active site modulates specificity in the interaction of O-acetylserine sulfhydrylase isozymes with serine acetyltransferase. Biochim. Biophys. Acta 2013, 1834, 169–181. [Google Scholar] [CrossRef] [PubMed]
- James, S.H.; Kimberlin, D.W. Advances in the prevention and treatment of congenital cytomegalovirus infection. Curr. Opin. Pediatr. 2016, 28, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Chemical Structure | Compound | IUPAC Name |
|---|---|---|
 | TH-EGO | (±)(E)-4-methyl-2-(((4-methyl-5-oxo-2,5-dihydrofuran-2-yl)oxy)methylene)-7-(thiophen-2-yl)-1,4-dihydrocyclopenta[b]indol-3(2H)-one |
 | EDOT-EGO | (±)(E)-7-(2,3-dihydrothieno[3,4-b][1,4]dioxin-5-yl)-4-methyl-2-(((4-methyl-5-oxo-2,5-dihydrofuran-2-yl)oxy)methylene)-1,4-dihydrocyclopenta[b]indol-3(2H)-one |
 | EGO-10 | (±)(E)-4-methyl-2-(((4-methyl-5-oxo-2,5-dihydrofuran-2-yl)oxy)methylene) -1,4-dihydrocyclopenta[b]indol-3(2H)-one |
 | GR24 | (±)(3aR,8bS,E)-3-((((R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl)oxy)methylene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one |
 | TH-ABC | 4-methyl-7-(thiophen-2-yl)-1,4-dihydrocyclopenta[b]indol-3(2H)-one |
 | EDOT-ABC | 7-(2,3-dihydrothieno[3,4-b][1,4]dioxin-5-yl)-4-methyl-1,4-dihydrocyclopenta[b]indol-3(2H)-one |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biolatti, M.; Blangetti, M.; D’Arrigo, G.; Spyrakis, F.; Cappello, P.; Albano, C.; Ravanini, P.; Landolfo, S.; De Andrea, M.; Prandi, C.; et al. Strigolactone Analogs Are Promising Antiviral Agents for the Treatment of Human Cytomegalovirus Infection. Microorganisms 2020, 8, 703. https://doi.org/10.3390/microorganisms8050703
Biolatti M, Blangetti M, D’Arrigo G, Spyrakis F, Cappello P, Albano C, Ravanini P, Landolfo S, De Andrea M, Prandi C, et al. Strigolactone Analogs Are Promising Antiviral Agents for the Treatment of Human Cytomegalovirus Infection. Microorganisms. 2020; 8(5):703. https://doi.org/10.3390/microorganisms8050703
Chicago/Turabian StyleBiolatti, Matteo, Marco Blangetti, Giulia D’Arrigo, Francesca Spyrakis, Paola Cappello, Camilla Albano, Paolo Ravanini, Santo Landolfo, Marco De Andrea, Cristina Prandi, and et al. 2020. "Strigolactone Analogs Are Promising Antiviral Agents for the Treatment of Human Cytomegalovirus Infection" Microorganisms 8, no. 5: 703. https://doi.org/10.3390/microorganisms8050703